Abstract
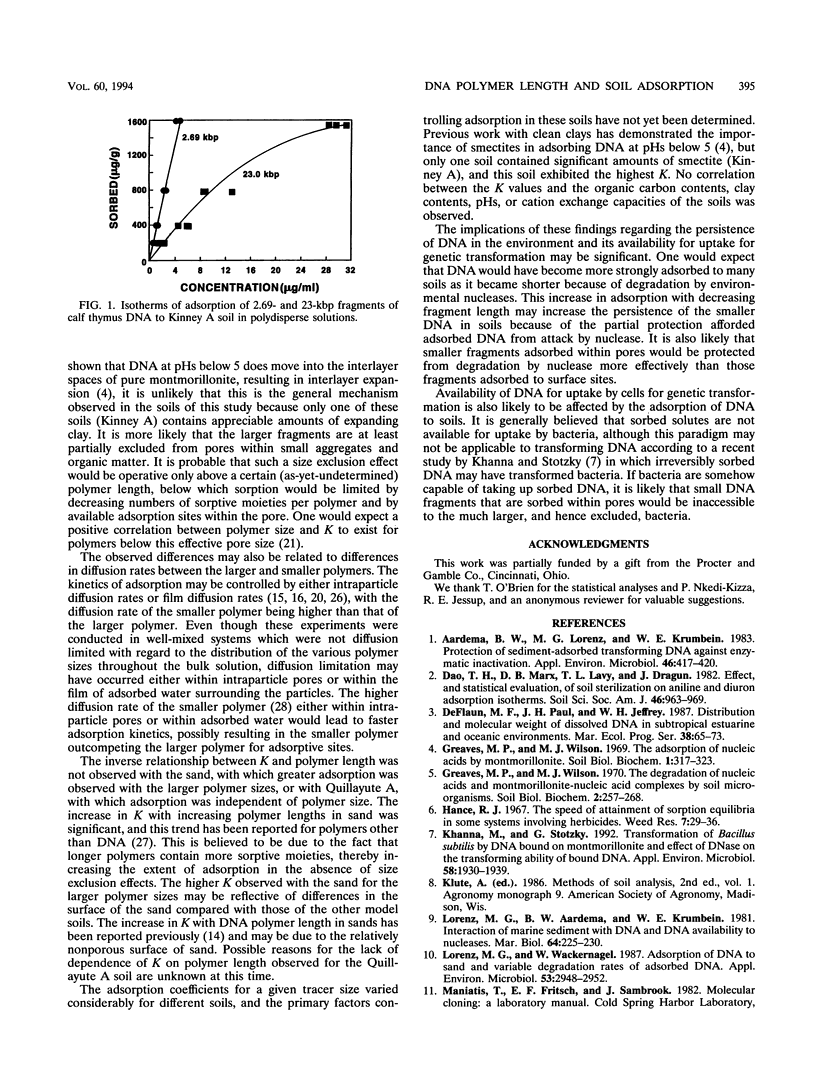
Three different DNA fragments ranging size from 2.69 kbp (1.75 MDa) to 23 kbp (14.95 MDa) were used as tracers to study the adsorption of polydisperse solutions of calf thymus DNA to eight model soils. The adsorption of the three tracers to all soils was described by the Freundlich adsorption model, with adsorption coefficients (K) ranging from 1.1 for acid-washed sand to over 300 for one soil. An inverse relationship between tracer size and K was observed with six of the eight soils, indicating that smaller fragments are sorbed preferentially versus larger fragments in these soils. No significant correlation between K and the organic carbon contents, clay contents, pHs, or cation exchange capacities of the model soils was observed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aardema B. W., Lorenz M. G., Krumbein W. E. Protection of sediment-adsorbed transforming DNA against enzymatic inactivation. Appl Environ Microbiol. 1983 Aug;46(2):417–420. doi: 10.1128/aem.46.2.417-420.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna M., Stotzky G. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl Environ Microbiol. 1992 Jun;58(6):1930–1939. doi: 10.1128/aem.58.6.1930-1939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl Environ Microbiol. 1987 Dec;53(12):2948–2952. doi: 10.1128/aem.53.12.2948-2952.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins E. J., Gordon M. P., Caceres O., Lurquin P. F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990 May;172(5):2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins E. J., Lurquin P. F. Duplication of a 2,4-dichlorophenoxyacetic acid monooxygenase gene in Alcaligenes eutrophus JMP134(pJP4). J Bacteriol. 1988 Dec;170(12):5669–5672. doi: 10.1128/jb.170.12.5669-5672.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol. 1991 Apr;57(4):1057–1061. doi: 10.1128/aem.57.4.1057-1061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangodkar U. M., Chapman P. J., Chakrabarty A. M. Cloning, physical mapping and expression of chromosomal genes specifying degradation of the herbicide 2,4,5-T by Pseudomonas cepacia AC1100. Gene. 1988 Nov 30;71(2):267–277. doi: 10.1016/0378-1119(88)90043-1. [DOI] [PubMed] [Google Scholar]


