Abstract
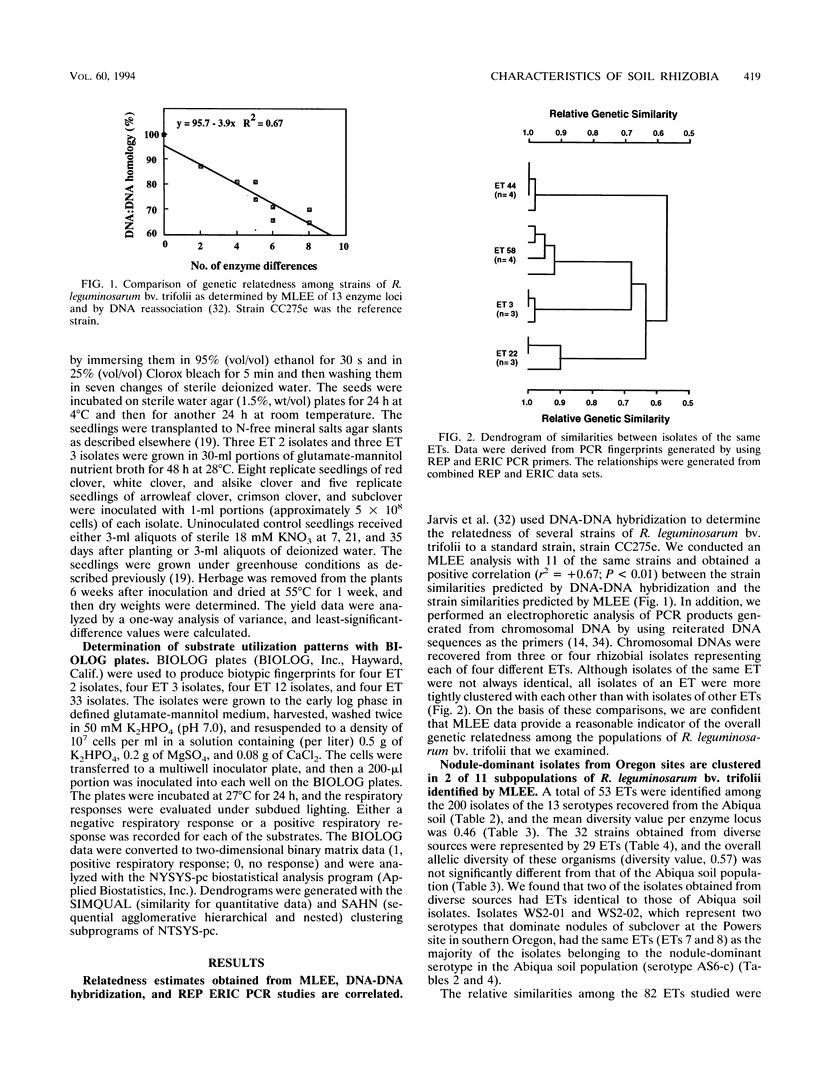
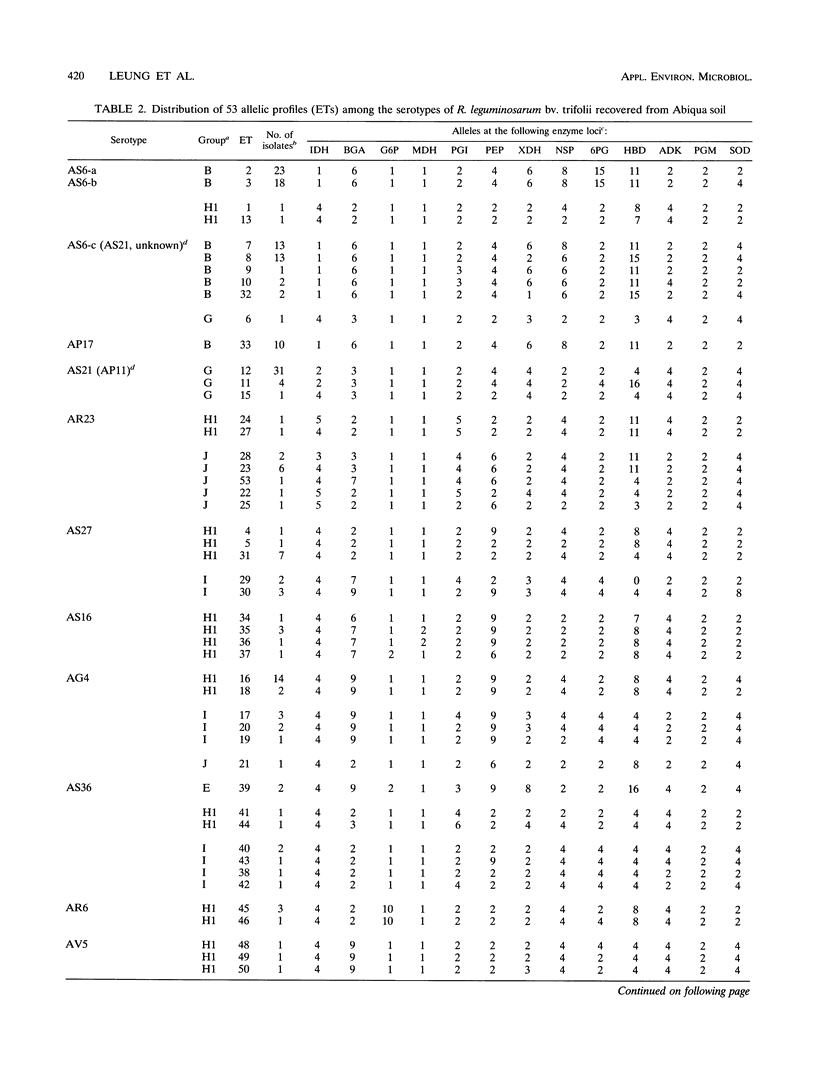
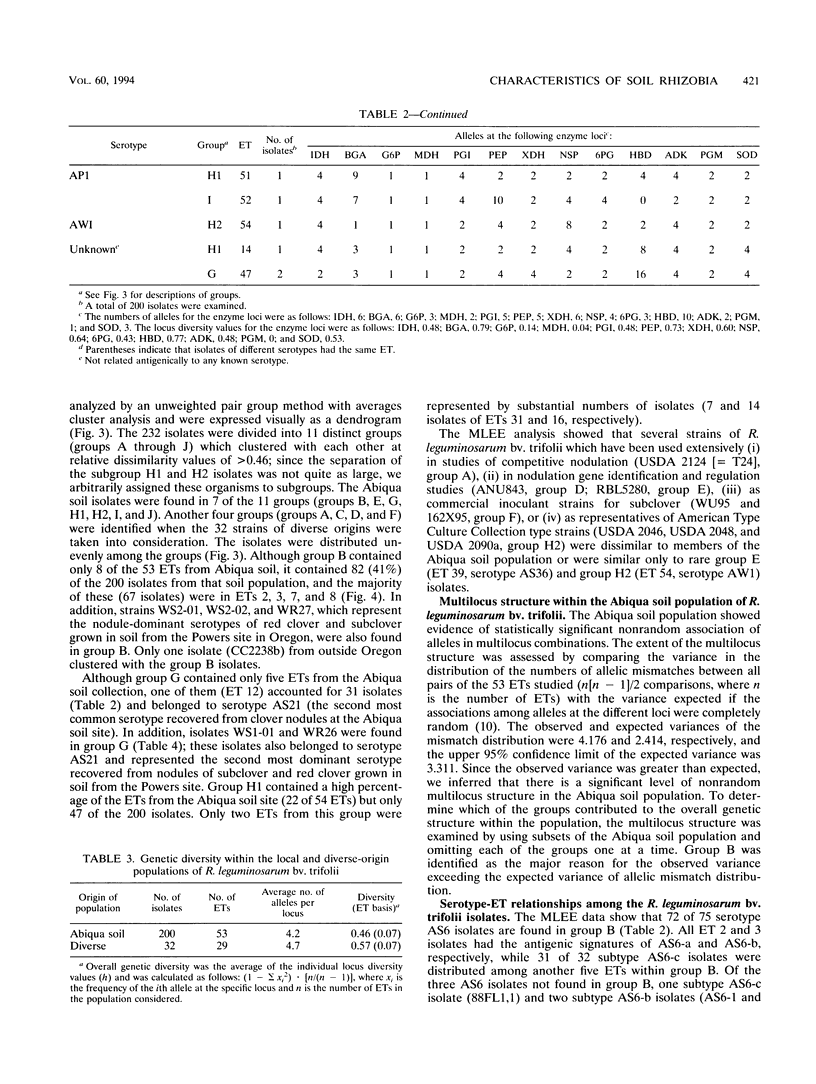
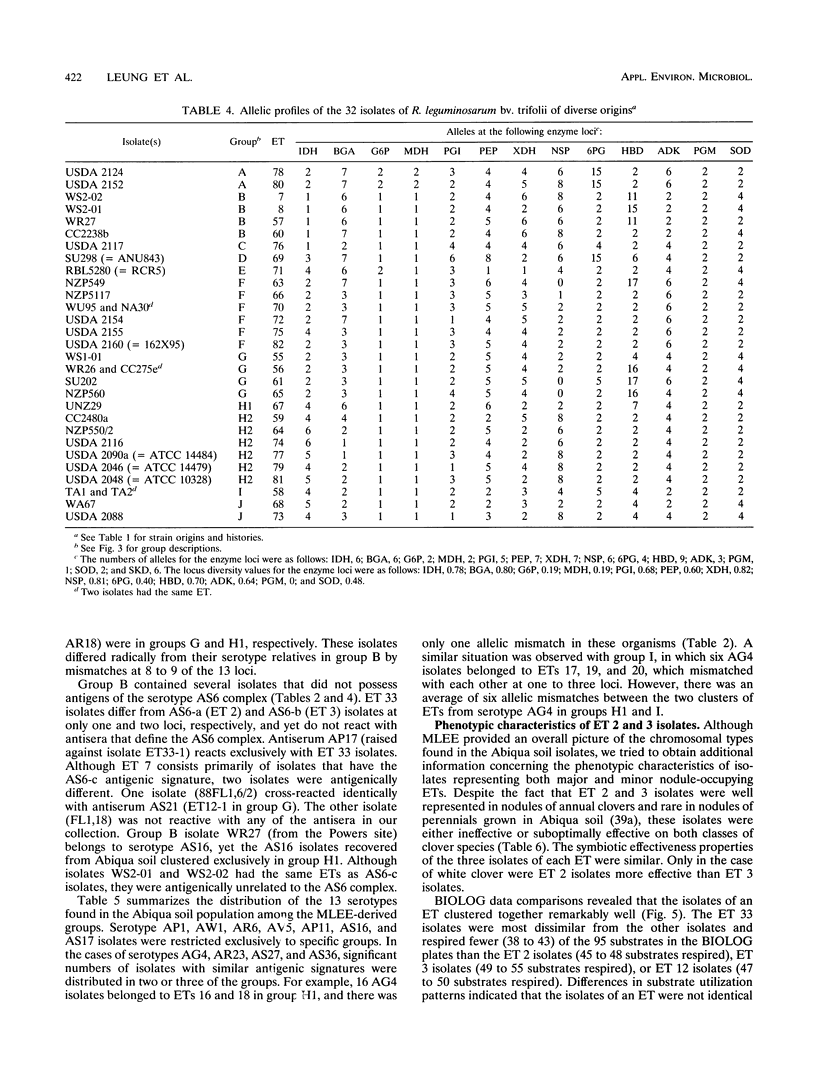
The relative genetic similarities of 200 isolates of Rhizobium leguminosarum bv. trifolii recovered from an Oregon soil were determined at 13 enzyme loci by multilocus enzyme electrophoresis (MLEE). These isolates represented 13 antigenically distinct serotypes recovered from nodules formed on various clover species. The MLEE-derived levels of relatedness among isolates of R. leguminosarum bv. trifolii were found to be in good agreement with the levels of relatedness established by using repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the PCR technique and with levels of relatedness from previously published DNA reassociation studies. BIOLOG substrate utilization patterns showed that isolates within an electrophoretic type (ET) were phenotypically more similar to each other than to isolates of other ETs. The soil isolates were represented by 53 ETs which could be clustered into seven groups (groups B, E, G, H1, H2, I, and J). Evidence for multilocus structure within the population was obtained, and group B was identified as the primary creator of the disequilibrium. Of 75 isolates belonging to the nodule-dominant serotype AS6 complex, 72 were found in group B. Isolates WS2-01 and WS2-02 representing nodule-dominant serotypes recovered from subclover grown at another Oregon site were also found in group B. Isolates representing the most numerous ETs in group B (ETs 2 and 3) were either suboptimally effective or completely ineffective at fixing nitrogen on six different clover species. Another four groups of isolates (groups A, C, D, and F) were identified when 32 strains of diverse origins were analyzed by MLEE and incorporated into the cluster analysis. Group A was most dissimilar in comparisons with other groups and contained strain USDA 2124 (T24), which produces trifolitoxin and has unique symbiotic characteristics.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayanaba A., Weiland K. D., Zablotowicz R. M. Evaluation of Diverse Antisera, Conjugates, and Support Media for Detecting Bradyrhizobium japonicum by Indirect Enzyme-Linked Immunosorbent Assay. Appl Environ Microbiol. 1986 Nov;52(5):1132–1138. doi: 10.1128/aem.52.5.1132-1138.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran P., Musser J. M., Helmuth R., Farmer J. J., 3rd, Frerichs W. M., Wachsmuth I. K., Ferris K., McWhorter A. C., Wells J. G., Cravioto A. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran P., Plock S. A., Smith N. H., Whittam T. S., Old D. C., Selander R. K. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J Gen Microbiol. 1991 Mar;137(3):601–606. doi: 10.1099/00221287-137-3-601. [DOI] [PubMed] [Google Scholar]
- Bottomley P. J., Dughri M. H. Population Size and Distribution of Rhizobium leguminosarum bv. trifolii in Relation to Total Soil Bacteria and Soil Depth. Appl Environ Microbiol. 1989 Apr;55(4):959–964. doi: 10.1128/aem.55.4.959-964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman F. J., Bezdicek D. F. Diversity within Serogroups of Rhizobium leguminosarum biovar viceae in the Palouse Region of Eastern Washington as Indicated by Plasmid Profiles, Intrinsic Antibiotic Resistance, and Topography. Appl Environ Microbiol. 1989 Jan;55(1):109–115. doi: 10.1128/aem.55.1.109-115.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. H., Feldman M. W., Nevo E. Multilocus Structure of Natural Populations of HORDEUM SPONTANEUM. Genetics. 1980 Oct;96(2):523–536. doi: 10.1093/genetics/96.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant D. A., Levin B. R., Orskov I., Orskov F., Svanborg Eden C., Selander R. K. Genetic diversity in relation to serotype in Escherichia coli. Infect Immun. 1985 Aug;49(2):407–413. doi: 10.1128/iai.49.2.407-413.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demezas D. H., Reardon T. B., Watson J. M., Gibson A. H. Genetic Diversity among Rhizobium leguminosarum bv. Trifolii Strains Revealed by Allozyme and Restriction Fragment Length Polymorphism Analyses. Appl Environ Microbiol. 1991 Dec;57(12):3489–3495. doi: 10.1128/aem.57.12.3489-3495.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle S. F., Bohlool B. B. Predominance of Fast-Growing Rhizobium japonicum in a Soybean Field in the People's Republic of China. Appl Environ Microbiol. 1985 Nov;50(5):1171–1176. doi: 10.1128/aem.50.5.1171-1176.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eardly B. D., Materon L. A., Smith N. H., Johnson D. A., Rumbaugh M. D., Selander R. K. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl Environ Microbiol. 1990 Jan;56(1):187–194. doi: 10.1128/aem.56.1.187-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedericks J. B., Hagedorn C., Vanscoyoc S. W. Isolation of Rhizobium leguminosarum (biovar trifolii) Strains from Ethiopian Soils and Symbiotic Effectiveness on African Annual Clover Species. Appl Environ Microbiol. 1990 Apr;56(4):1087–1092. doi: 10.1128/aem.56.4.1087-1092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann J., Wollum A. G. Simplified Enzyme-Linked Immunosorbent Assay for Routine Identification of Rhizobium japonicum Antigens. Appl Environ Microbiol. 1985 Apr;49(4):1010–1013. doi: 10.1128/aem.49.4.1010-1013.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour M. N., Whittam T. S., Kilian M., Selander R. K. Genetic relationships among the oral streptococci. J Bacteriol. 1987 Nov;169(11):5247–5257. doi: 10.1128/jb.169.11.5247-5257.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A. A. Serologic characteristics of certain root-nodule bacteria of legumes. Antonie Van Leeuwenhoek. 1966;32(4):410–418. doi: 10.1007/BF02097492. [DOI] [PubMed] [Google Scholar]
- Istock C. A., Duncan K. E., Ferguson N., Zhou X. Sexuality in a natural population of bacteria--Bacillus subtilis challenges the clonal paradigm. Mol Ecol. 1992 Aug;1(2):95–103. doi: 10.1111/j.1365-294x.1992.tb00161.x. [DOI] [PubMed] [Google Scholar]
- Judd A. K., Schneider M., Sadowsky M. J., de Bruijn F. J. Use of repetitive sequences and the polymerase chain reaction technique to classify genetically related Bradyrhizobium japonicum serocluster 123 strains. Appl Environ Microbiol. 1993 Jun;59(6):1702–1708. doi: 10.1128/aem.59.6.1702-1708.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamicker B. J., Brill W. J. Identification of Bradyrhizobium japonicum Nodule Isolates from Wisconsin Soybean Farms. Appl Environ Microbiol. 1986 Mar;51(3):487–492. doi: 10.1128/aem.51.3.487-492.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang U. G., Somasegaran P., Hoben H. J., Bohlool B. B. Symbiotic Potential, Competitiveness, and Serological Properties of Bradyrhizobium japonicum Indigenous to Korean Soils. Appl Environ Microbiol. 1991 Apr;57(4):1038–1045. doi: 10.1128/aem.57.4.1038-1045.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K., Yap K., Dashti N., Bottomley P. J. Serological and Ecological Characteristics of a Nodule-Dominant Serotype from an Indigenous Soil Population of Rhizobium leguminosarum bv. trifolii. Appl Environ Microbiol. 1994 Feb;60(2):408–415. doi: 10.1128/aem.60.2.408-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Romero E., Segovia L., Mercante F. M., Franco A. A., Graham P., Pardo M. A. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int J Syst Bacteriol. 1991 Jul;41(3):417–426. doi: 10.1099/00207713-41-3-417. [DOI] [PubMed] [Google Scholar]
- Noel K. D., Brill W. J. Diversity and Dynamics of Indigenous Rhizobium japonicum Populations. Appl Environ Microbiol. 1980 Nov;40(5):931–938. doi: 10.1128/aem.40.5.931-938.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Evidence for clonal population structure in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(1):198–201. doi: 10.1073/pnas.81.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Whittam T. S., Caugant D. A., Selander R. K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983 Sep;129(9):2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- Pinero D., Martinez E., Selander R. K. Genetic diversity and relationships among isolates of Rhizobium leguminosarum biovar phaseoli. Appl Environ Microbiol. 1988 Nov;54(11):2825–2832. doi: 10.1128/aem.54.11.2825-2832.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Cregan P. B., Keyser H. H. DNA Hybridization Probe for Use in Determining Restricted Nodulation among Bradyrhizobium japonicum Serocluster 123 Field Isolates. Appl Environ Microbiol. 1990 Jun;56(6):1768–1774. doi: 10.1128/aem.56.6.1768-1774.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Tully R. E., Cregan P. B., Keyser H. H. Genetic Diversity in Bradyrhizobium japonicum Serogroup 123 and Its Relation to Genotype-Specific Nodulation of Soybean. Appl Environ Microbiol. 1987 Nov;53(11):2624–2630. doi: 10.1128/aem.53.11.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. L., Bakole R. O., Bohlool B. B. Fluorescent-antibody approach to study of rhizobia in soil. J Bacteriol. 1968 Jun;95(6):1987–1992. doi: 10.1128/jb.95.6.1987-1992.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. L., Zidwick M. J., Abebe H. M. Bradyrhizobium japonicum Serocluster 123 and Diversity among Member Isolates. Appl Environ Microbiol. 1986 Jun;51(6):1212–1215. doi: 10.1128/aem.51.6.1212-1215.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. R., Gibson A. H., Dudman W. F., Watson J. M. Evidence for genetic exchange and recombination of Rhizobium symbiotic plasmids in a soil population. Appl Environ Microbiol. 1987 Dec;53(12):2942–2947. doi: 10.1128/aem.53.12.2942-2947.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinghamer E. A., Belkengren R. P. Inhibition of rhizobia by a strain of Rhizobeium trifolii: some properties of the antibiotic and of the strain. Arch Mikrobiol. 1968;64(2):130–145. doi: 10.1007/BF00406972. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., McKinney R. M., Whittam T. S., Bibb W. F., Brenner D. J., Nolte F. S., Pattison P. E. Genetic structure of populations of Legionella pneumophila. J Bacteriol. 1985 Sep;163(3):1021–1037. doi: 10.1128/jb.163.3.1021-1037.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza V., Nguyen T. T., Hudson R. R., Piñero D., Lenski R. E. Hierarchical analysis of linkage disequilibrium in Rhizobium populations: evidence for sex? Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8389–8393. doi: 10.1073/pnas.89.17.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett E. W., Barta T. M. Trifolitoxin Production and Nodulation Are Necessary for the Expression of Superior Nodulation Competitiveness by Rhizobium leguminosarum bv. trifolii Strain T24 on Clover. Plant Physiol. 1987 Oct;85(2):335–342. doi: 10.1104/pp.85.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Wilson R. A. Genetic relationships among pathogenic strains of avian Escherichia coli. Infect Immun. 1988 Sep;56(9):2458–2466. doi: 10.1128/iai.56.9.2458-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. P., Demetriou L., Apte R. G. Rhizobium Population Genetics: Enzyme Polymorphism in Rhizobium leguminosarum from Plants and Soil in a Pea Crop. Appl Environ Microbiol. 1987 Feb;53(2):397–402. doi: 10.1128/aem.53.2.397-402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992 Jul;58(7):2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]


