Abstract
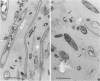
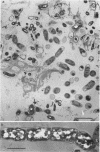
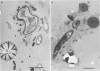
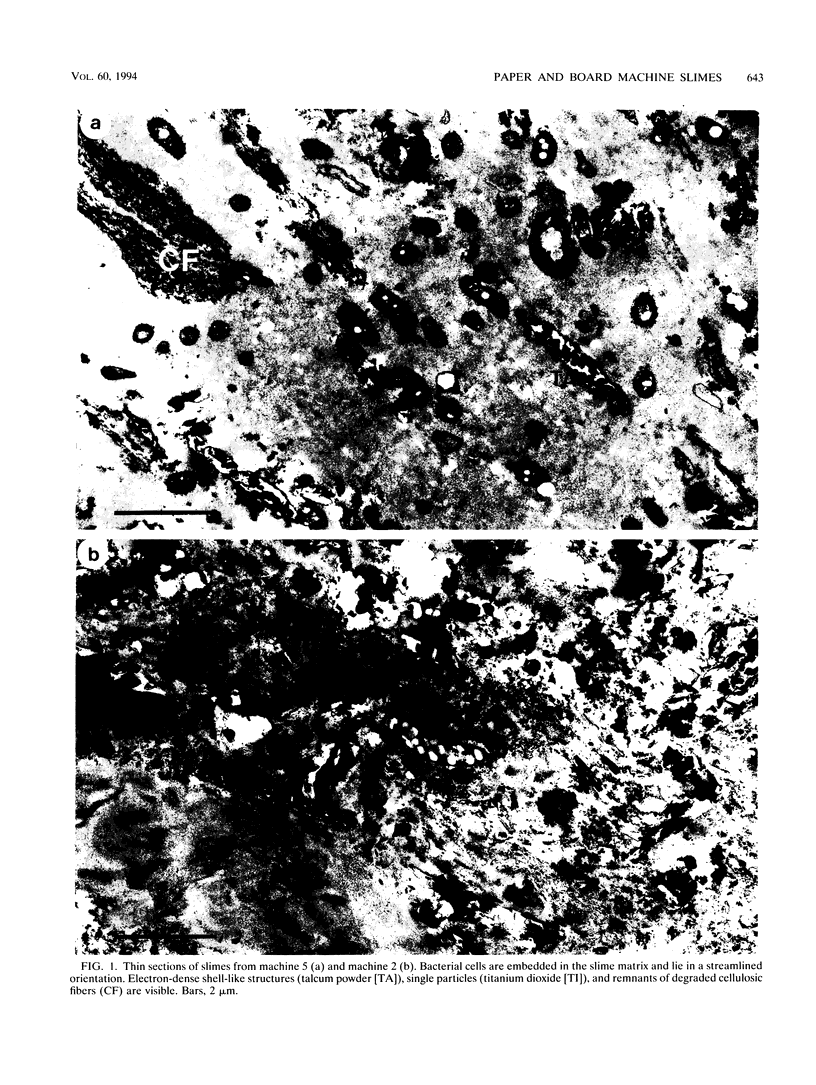
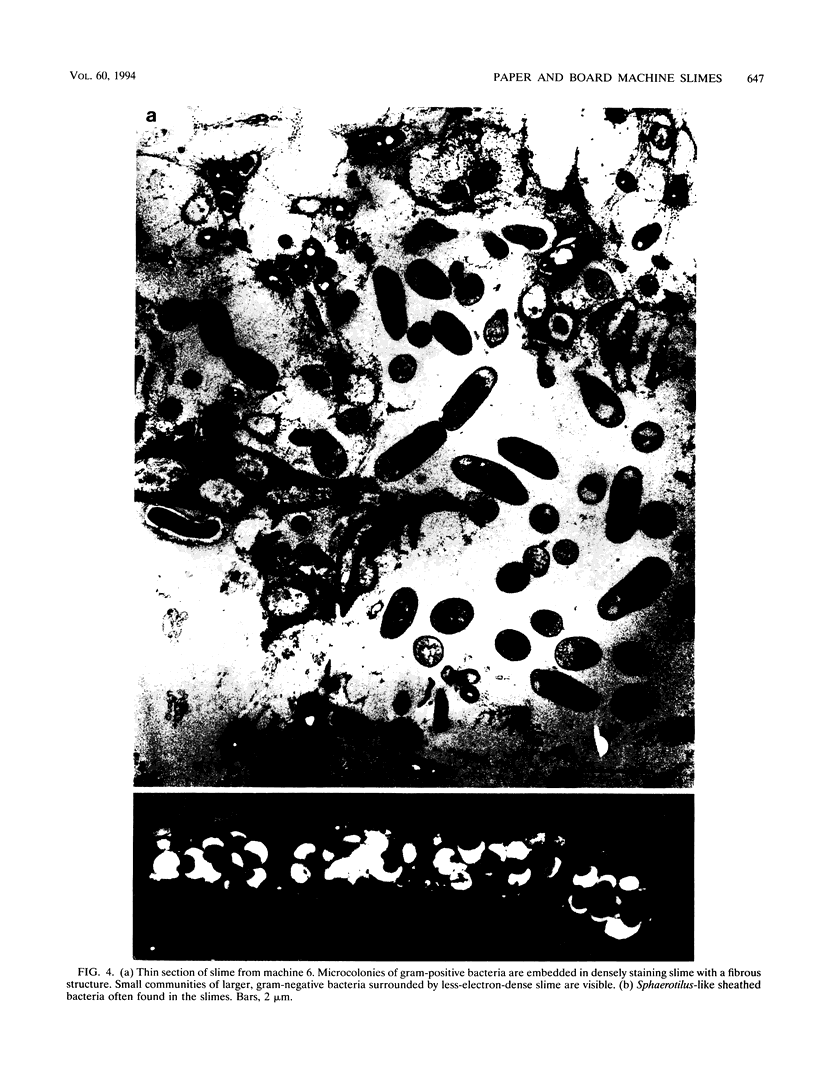
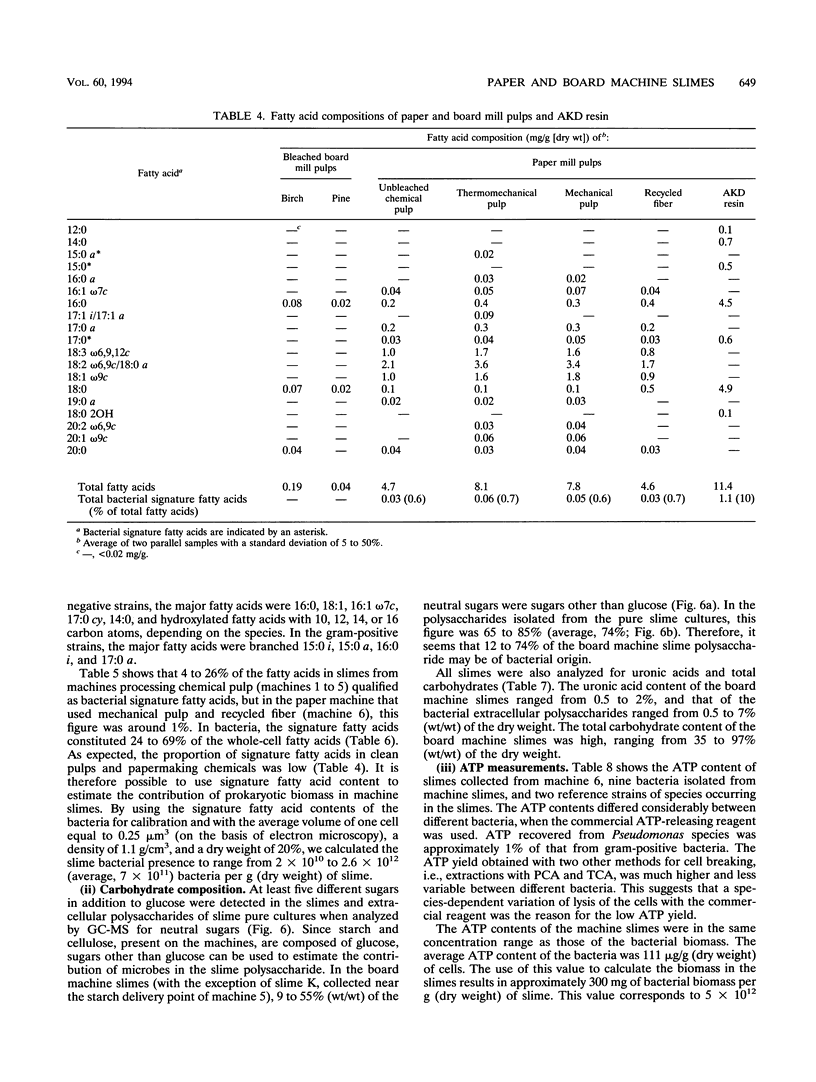
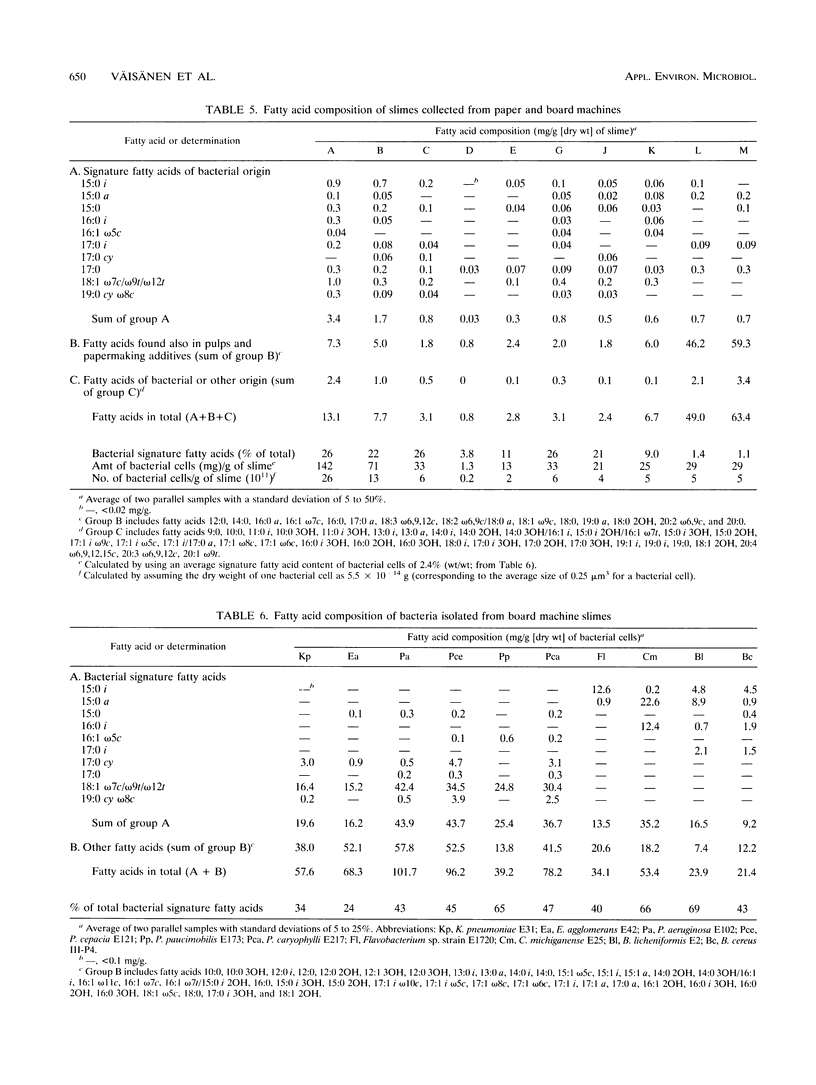
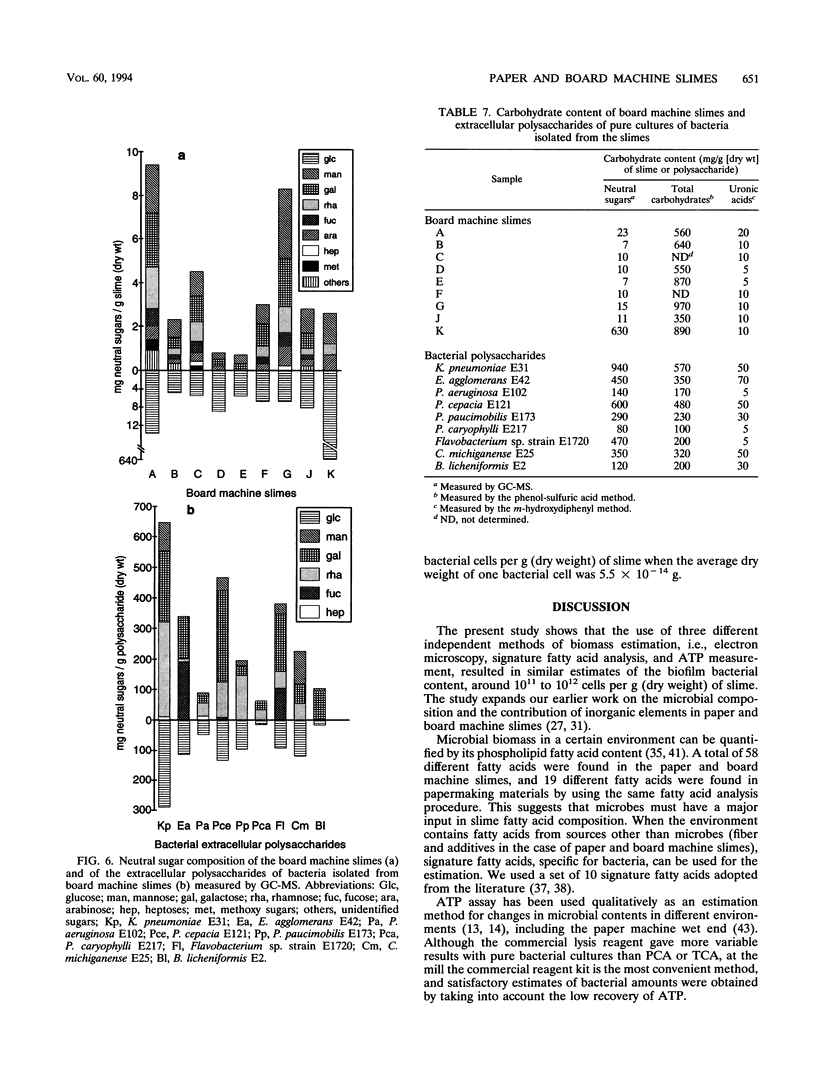
Biological slimes (biofilms) collected from the wet end of paper and board machines were examined by electron microscopy and analyzed for fatty acid composition, neutral sugar composition, and ATP. Electron microscopy revealed minuscule prokaryotic organisms (diameter, 0.2 to 0.4 μm). Larger cells morphologically resembling Sphaerotilus and Leptothrix spp. were found in slimes from machines using recycled fiber or unbleached pulp. The bacteria were embedded in a slimy matrix and often contained reserve materials microscopically resembling poly-β-hydroxybutyrate and glycogen. Fatty acid analysis of the slimes revealed bacterial signature fatty acids in concentrations equivalent to the presence of 2 × 1010 to 2.6 × 1012 (average, 7 × 1011) bacterial cells (live and dead) per g (dry weight) of slime. The slimes contained several known components of bacterial polysaccharides in addition to glucose, indicating that the slime body consisted of bacterial polysaccharides. The slimes contained uronic acids equivalent to a binding capacity of 12.5 to 50 μmol of divalent cations per g (dry weight) of slime. The uronic acid-containing polysaccharides may be responsible for the accumulation of heavy metals in the slime. Calculation of the ATP contents of the slimes resulted in an estimate of 5 × 1012 cells per g (dry weight) of slime when calibrated with pure bacterial cultures isolated from the slimes. From electron micrographs, an estimate ranging from 1 × 1010 to 1.5 × 1012 (average, 4 × 1011) cells per g (dry weight) of slime was obtained.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumenkrantz N., Asboe-Hansen G. New method for quantitative determination of uronic acids. Anal Biochem. 1973 Aug;54(2):484–489. doi: 10.1016/0003-2697(73)90377-1. [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Cheng K. J., Geesey G. G., Ladd T. I., Nickel J. C., Dasgupta M., Marrie T. J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Eighmy T. T., Maratea D., Bishop P. L. Electron microscopic examination of wastewater biofilm formation and structural components. Appl Environ Microbiol. 1983 Jun;45(6):1921–1931. doi: 10.1128/aem.45.6.1921-1931.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner N. E., Balkwill D. L., Bishop P. L. Light and electron microscopic studies of microorganisms growing in rotating biological contactor biofilms. Appl Environ Microbiol. 1983 May;45(5):1659–1669. doi: 10.1128/aem.45.5.1659-1669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeChevallier M. W., Cawthon C. D., Lee R. G. Inactivation of biofilm bacteria. Appl Environ Microbiol. 1988 Oct;54(10):2492–2499. doi: 10.1128/aem.54.10.2492-2499.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian F. A., Jarman T. R., Righelato R. C. Biosynthesis of exopolysaccharide by Pseudomonas aeruginosa. J Bacteriol. 1978 May;134(2):418–422. doi: 10.1128/jb.134.2.418-422.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway H. F., Kelly A., Justice C., Olson B. H. Microbial fouling of reverse-osmosis membranes used in advanced wastewater treatment technology: chemical, bacteriological, and ultrastructural analyses. Appl Environ Microbiol. 1983 Mar;45(3):1066–1084. doi: 10.1128/aem.45.3.1066-1084.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively J. M. Inclusion bodies of prokaryotes. Annu Rev Microbiol. 1974;28(0):167–187. doi: 10.1146/annurev.mi.28.100174.001123. [DOI] [PubMed] [Google Scholar]
- Tunlid A., Hoitink H. A., Low C., White D. C. Characterization of bacteria that suppress rhizoctonia damping-off in bark compost media by analysis of Fatty Acid biomarkers. Appl Environ Microbiol. 1989 Jun;55(6):1368–1374. doi: 10.1128/aem.55.6.1368-1374.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väisänen O. M., Mentu J., Salkinoja-Salonen M. S. Bacteria in food packaging paper and board. J Appl Bacteriol. 1991 Aug;71(2):130–133. doi: 10.1111/j.1365-2672.1991.tb02967.x. [DOI] [PubMed] [Google Scholar]