Abstract
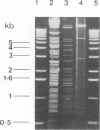
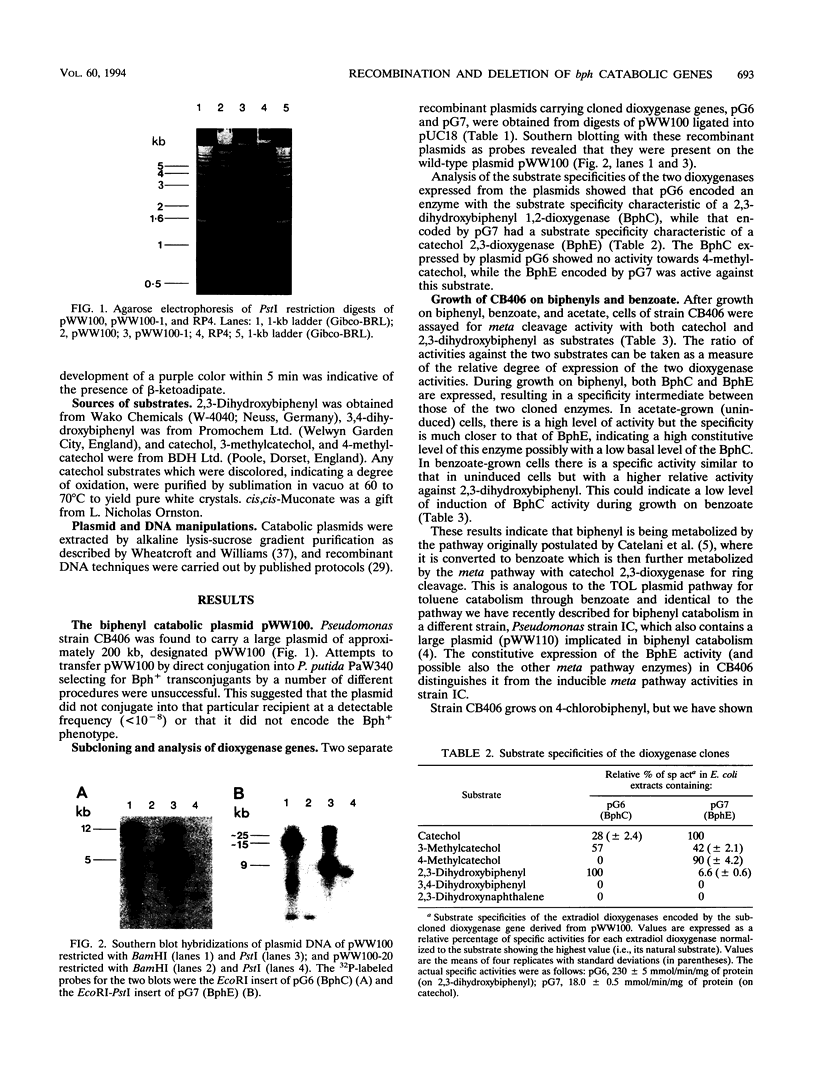
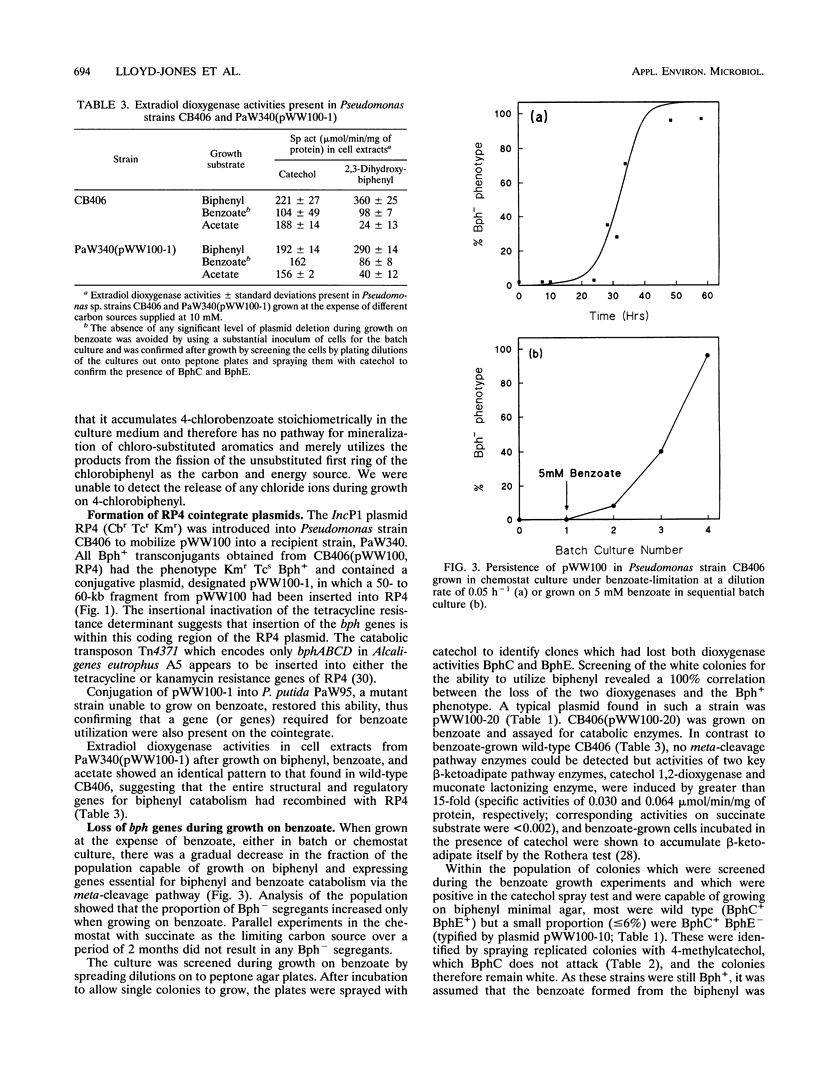
Pseudomonas sp. strain CB406 was isolated from polychlorinated biphenyl-contaminated soil and harbors a nontransmissible plasmid, pWW100, of approximately 200 kb which carries the genes required for biphenyl and 4-chlorobiphenyl catabolism. The catabolic phenotype was mobilized following the construction in vivo of a cointegrate plasmid containing functional upper and lower biphenyl operons inserted into the broad-host-range R plasmid RP4. The Bph+ phenotype carried by pWW100 was stable in nonselective media but was unstable during growth on benzoate, where the sequential selection of two species of bph deletion derivatives occurs at high frequency. This mirrors observations made with TOL plasmids (encoding toluene and xylene catabolism) grown under similar conditions. Subcloning of dioxygenase genes involved in biphenyl catabolism confirmed the localization of the bph genes on the wild-type plasmid and the RP4 cointegrate plasmid.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad D., Massé R., Sylvestre M. Cloning and expression of genes involved in 4-chlorobiphenyl transformation by Pseudomonas testosteroni: homology to polychlorobiphenyl-degrading genes in other bacteria. Gene. 1990 Jan 31;86(1):53–61. doi: 10.1016/0378-1119(90)90113-6. [DOI] [PubMed] [Google Scholar]
- Assinder S. J., Williams P. A. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv Microb Physiol. 1990;31:1–69. doi: 10.1016/s0065-2911(08)60119-8. [DOI] [PubMed] [Google Scholar]
- Bayley S. A., Duggleby C. J., Worsey M. J., Williams P. A., Hardy K. G., Broda P. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol Gen Genet. 1977 Jul 20;154(2):203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- Catelani D., Sorlini C., Treccani V. The metabolism of biphenyl by Pseudomonas putida. Experientia. 1971 Oct 15;27(10):1173–1174. doi: 10.1007/BF02286908. [DOI] [PubMed] [Google Scholar]
- Duetz W. A., Winson M. K., van Andel J. G., Williams P. A. Mathematical analysis of catabolic function loss in a population of Pseudomonas putida mt-2 during non-limited growth on benzoate. J Gen Microbiol. 1991 Jun;137(6):1363–1368. doi: 10.1099/00221287-137-6-1363. [DOI] [PubMed] [Google Scholar]
- Duetz W. A., van Andel J. G. Stability of TOL plasmid pWW0 in Pseudomonas putida mt-2 under non-selective conditions in continuous culture. J Gen Microbiol. 1991 Jun;137(6):1369–1374. doi: 10.1099/00221287-137-6-1369. [DOI] [PubMed] [Google Scholar]
- Erickson B. D., Mondello F. J. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992 May;174(9):2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Williams P. A. Construction of a partial diploid for the degradative pathway encoded by the TOL plasmid (pWWO) from Pseudomonas putida mt-2: evidence for the positive nature of the regulation by the xyIR gene. Mol Gen Genet. 1980 Jan;177(2):321–328. doi: 10.1007/BF00267445. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Arimura N., Miyazaki T. Nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene of Pseudomonas pseudoalcaligenes. J Bacteriol. 1987 Jan;169(1):427–429. doi: 10.1128/jb.169.1.427-429.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Chakrabarty A. M. Involvement of plasmids in total degradation of chlorinated biphenyls. Appl Environ Microbiol. 1982 Sep;44(3):619–626. doi: 10.1128/aem.44.3.619-626.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Hayase N., Taira K., Tomizuka N. Molecular relationship of chromosomal genes encoding biphenyl/polychlorinated biphenyl catabolism: some soil bacteria possess a highly conserved bph operon. J Bacteriol. 1989 Oct;171(10):5467–5472. doi: 10.1128/jb.171.10.5467-5472.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Miyazaki T. Cloning of a gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J Bacteriol. 1986 May;166(2):392–398. doi: 10.1128/jb.166.2.392-398.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper S. W., Dockendorff T. C., Sayler G. S. Characteristics and restriction analysis of the 4-chlorobiphenyl catabolic plasmid, pSS50. Appl Environ Microbiol. 1989 May;55(5):1286–1288. doi: 10.1128/aem.55.5.1286-1288.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeenes D. J., Williams P. A. Excision and integration of degradative pathway genes from TOL plasmid pWW0. J Bacteriol. 1982 Apr;150(1):188–194. doi: 10.1128/jb.150.1.188-194.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Keil H., Keil S., Pickup R. W., Williams P. A. Evolutionary conservation of genes coding for meta pathway enzymes within TOL plasmids pWW0 and pWW53. J Bacteriol. 1985 Nov;164(2):887–895. doi: 10.1128/jb.164.2.887-895.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbara K., Hashimoto T., Fukuda M., Koana T., Takagi M., Oishi M., Yano K. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989 May;171(5):2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhm A. E., Stolz A., Ngai K. L., Knackmuss H. J. Purification and characterization of a 1,2-dihydroxynaphthalene dioxygenase from a bacterium that degrades naphthalenesulfonic acids. J Bacteriol. 1991 Jun;173(12):3795–3802. doi: 10.1128/jb.173.12.3795-3802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Meulien P., Downing R. G., Broda P. Excision of the 40kb segment of the TOL plasmid from Pseudomonas putida mt-2 involves direct repeats. Mol Gen Genet. 1981;184(1):97–101. doi: 10.1007/BF00271202. [DOI] [PubMed] [Google Scholar]
- Mondello F. J. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J Bacteriol. 1989 Mar;171(3):1725–1732. doi: 10.1128/jb.171.3.1725-1732.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu C., Ng J., Singh R., Straus N., Wyndham C. Chlorobenzoate catabolic transposon Tn5271 is a composite class I element with flanking class II insertion sequences. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8312–8316. doi: 10.1073/pnas.88.19.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne D. J., Pickup R. W., Williams P. A. The presence of two complete homologous meta pathway operons on TOL plasmid pWW53. J Gen Microbiol. 1988 Nov;134(11):2965–2975. doi: 10.1099/00221287-134-11-2965. [DOI] [PubMed] [Google Scholar]
- Rothera A. C. Note on the sodium nitro-prusside reaction for acetone. J Physiol. 1908 Dec 15;37(5-6):491–494. doi: 10.1113/jphysiol.1908.sp001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael D., Kreps S., Mergeay M. Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J Bacteriol. 1993 Mar;175(6):1674–1681. doi: 10.1128/jb.175.6.1674-1681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens G. M., Dalton H. The effect of lipophilic weak acids on the segregational stability of TOL plasmids in Pseudomonas putida. J Gen Microbiol. 1987 Jul;133(7):1891–1899. doi: 10.1099/00221287-133-7-1891. [DOI] [PubMed] [Google Scholar]
- Taira K., Hayase N., Arimura N., Yamashita S., Miyazaki T., Furukawa K. Cloning and nucleotide sequence of the 2,3-dihydroxybiphenyl dioxygenase gene from the PCB-degrading strain of Pseudomonas paucimobilis Q1. Biochemistry. 1988 May 31;27(11):3990–3996. doi: 10.1021/bi00411a015. [DOI] [PubMed] [Google Scholar]
- Taira K., Hirose J., Hayashida S., Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992 Mar 5;267(7):4844–4853. [PubMed] [Google Scholar]
- Tsuda M., Iino T. Naphthalene degrading genes on plasmid NAH7 are on a defective transposon. Mol Gen Genet. 1990 Aug;223(1):33–39. doi: 10.1007/BF00315794. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Minegishi K., Iino T. Toluene transposons Tn4651 and Tn4653 are class II transposons. J Bacteriol. 1989 Mar;171(3):1386–1393. doi: 10.1128/jb.171.3.1386-1393.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatcroft R., Williams P. A. Rapid methods for the study of both stable and unstable plasmids in Pseudomonas. J Gen Microbiol. 1981 Jun;124(2):433–437. doi: 10.1099/00221287-124-2-433. [DOI] [PubMed] [Google Scholar]
- Williams P. A., Taylor S. D., Gibb L. E. Loss of the toluene-xylene catabolic genes of TOL plasmid pWW0 during growth of Pseudomonas putida on benzoate is due to a selective growth advantage of 'cured' segregants. J Gen Microbiol. 1988 Jul;134(7):2039–2048. doi: 10.1099/00221287-134-7-2039. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yates J. R., Mondello F. J. Sequence similarities in the genes encoding polychlorinated biphenyl degradation by Pseudomonas strain LB400 and Alcaligenes eutrophus H850. J Bacteriol. 1989 Mar;171(3):1733–1735. doi: 10.1128/jb.171.3.1733-1735.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. R., Zehnder A. J., de Vos W. M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J Bacteriol. 1991 Nov;173(22):7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]