Abstract
Results of electrophoretic surveys have suggested that hemoglobin polymorphism may be maintained by balancing selection in natural populations of house mice, Mus musculus. Here we report a survey of nucleotide variation in the adult globin genes of house mice from South America. We surveyed nucleotide polymorphism in two closely linked α-globin paralogs and two closely linked β-globin paralogs to test whether patterns of variation are consistent with a model of long-term balancing selection. Surprisingly high levels of nucleotide polymorphism at the two β-globin paralogs were attributable to the segregation of two highly divergent haplotypes, Hbbs (which carries two identical β-globin paralogs) and Hbbd (which carries two functionally divergent β-globin paralogs). Interparalog gene conversion on the Hbbs haplotype has produced a highly unusual situation in which the two paralogs are more similar to one another than either one is to its allelic counterpart on the Hbbd haplotype. Levels of nucleotide polymorphism and linkage disequilibrium at the two β-globin paralogs suggest a complex history of diversity-enhancing selection that may be responsible for long-term maintenance of alternative protein alleles. The alternative two-locus β-globin haplotypes are associated with pronounced differences in intraerythrocyte glutathione and nitric oxide metabolism, suggesting a possible mechanism for selection on hemoglobin function.
NATURAL populations of house mice, Mus musculus, exhibit high levels of hemoglobin polymorphism and evidence suggests that allelic variation in the β-chain subunit polypeptides is maintained by some form of balancing selection (reviewed in Berry 1978). In Mus, as in other gnathostome vertebrates, the α- and β-chain subunits of the tetrameric hemoglobin protein are encoded by different sets of duplicated genes that are located on different chromosomes. In M. musculus, the α-chain subunits of adult hemoglobin are encoded by two duplicated genes that are separated by 12.0 kb on chromosome 11, and the β-chain subunits are encoded by two duplicated genes that are separated by 12.6 kb on chromosome 7 (Figure 1). A number of distinct two-locus α-globin haplotypes have been described among different inbred strains of M. musculus (Erhart et al. 1987). Some strains carry two-locus α-globin haplotypes in which the two paralogs encode identical polypeptides, while other strains carry haplotypes in which the two paralogs are distinguished by 1–5 amino acid substitutions (Hilse and Popp 1968; Nishioka and Leder 1979; Whitney et al. 1979, 1985; Leder et al. 1980; Popp et al. 1982; Erhart et al. 1987). Studies of nucleotide variation and amino acid variation in the adult β-globin genes of inbred strains have also revealed a number of highly distinct two-locus haplotypes (Erhart et al. 1985). As with the two α-globin paralogs, some strains carry two-locus β-globin haplotypes in which the two paralogs encode identical polypeptides, while other strains carry haplotypes in which the two paralogs are distinguished by 9–11 amino acid substitutions (Hutton et al. 1962; Popp 1973; Gilman 1976; Leder et al. 1980; Erhart et al. 1985).
Figure 1.—
Genomic structure of the α- and β-globin gene families of M. musculus. Location of the two adult α-globin paralogs, HBA-T1 and HBA-T2, on chromosome 11 (top). Location of the two adult β-globin paralogs, HBB-T1 and HBB-T2, on chromosome 7 (bottom).
Electrophoretic surveys of hemoglobin polymorphism among inbred strains of house mice have revealed two main β-globin variants, HBB-S and HBB-D. Strains that carry the Hbbs haplotype (e.g., C57BL/6J) possess two β-globin paralogs that encode identical polypeptides. β-Globins produced by Hbbs mice migrate during electrophoresis as a dimer with α-globin and produce a single band (hence the ‘S’ designation; Whitney 1978). Strains that carry the Hbbd haplotype (e.g., BALBc/ByJ) possess two β-globin paralogs that encode structurally distinct polypeptides. β-Globins produced by Hbbd mice migrate during electrophoresis as a dimer with α-globin and produce double bands (hence the ‘D’ designation). Electrophoretic surveys of natural populations of house mice have revealed that the HBB-S and HBB-D electromorphs are maintained at intermediate frequencies in geographically disparate populations across the species' range, a pattern that is not paralleled at other unlinked loci (Petras 1967; Selander and Yang 1969; Selander et al. 1969a,b; Berry and Murphy 1970; Myers 1974; Berry and Jakobson 1975; Berry and Peters 1975, 1977, 1981; Berry et al. 1978; Sage 1981; Sage et al. 1986). In both Old and New World populations, the β-globin genes consistently exhibit the highest levels of heterozygosity in multilocus surveys of protein polymorphism (Selander and Yang 1969; Selander et al. 1969b) and a number of researchers have suggested that the allelic variation is maintained by overdominance of fitness or some other form of balancing selection (Selander et al. 1969b; Berry and Murphy 1970; Wheeler and Selander 1972; Myers 1974; Berry and Peters 1975, 1977; Berry 1978; Berry et al. 1978; Petras and Topping 1983). A number of studies have documented pronounced shifts in β-globin allele frequencies between seasons and between age classes (Berry and Murphy 1970; Bellamy et al. 1973; Myers 1974; Berry and Peters 1975, 1977; Berry 1978; Berry et al. 1978; Petras and Topping 1983), suggesting that temporally varying selection may play a role in maintaining the polymorphism. Some authors have also suggested that patterns of geographic variation in β-globin allele frequencies may reflect local adaptation to different climatic regimes (Selander et al. 1969b; Wheeler and Selander 1972).
The various α- and β-chain hemoglobin isoforms of house mice are characterized by different oxygen-binding affinities (Newton and Peters 1983; D'surney and Popp 1992), and allelic variation in β-chain cysteine content exerts a strong influence on intraerythrocyte glutathione and nitric oxide (NO) metabolism (Giustarini et al. 2006; Hempe et al. 2007). Despite the well-documented functional differences between the s- and d-type β-globin alleles and the suggestive evidence for balancing selection, a mechanistic link between allelic variation in hemoglobin function and fitness-related physiological performance has yet to be established.
Since modifications of hemoglobin function often play a key role in adaptation to high-altitude hypoxia (Perutz 1983; Monge and León-Velarde 1991; Poyart et al. 1992; Storz 2007), surveys of hemoglobin variation between high- and low-altitude populations may be especially informative about the role of selection in shaping patterns of functional variation at globin genes. In high-altitude environments, the reduced partial pressure of oxygen results in reduced pulmonary oxygen loading such that arterial blood may not carry a sufficient supply of oxygen to the cells of respiring tissues (Turek et al. 1973; Bencowitz et al. 1982; Bouverot 1985). This reduced level of tissue oxygenation can impose severe constraints on aerobic metabolism (Hayes 1989). One of the most important mechanisms to compensate for a reduced arterial partial pressure of oxygen involves increasing the circulatory conductance of oxygen in the bloodstream. This can be accomplished by increasing hemoglobin concentration in the blood or by changing the oxygen-binding affinity of hemoglobin (Storz 2007). In mammals, the former mechanism appears to be more important in the acclimation response to hypoxia in species that are native to lowland environments. By contrast, the latter mechanism appears to be more important in species that are native to high-altitude environments and are therefore genetically adapted to chronic hypoxia (Bullard 1972; Lenfant 1973; Monge and León-Velarde 1991; Hochachka and Somero 2002). It is unclear what mechanisms of adaptation or physiological acclimation may be important in species that have only recently colonized high-altitude environments.
M. musculus is a predominantly lowland, palearctic species that has recently colonized high-altitude environments in the Andes of South America. House mice colonized the New World within the past several hundred years in conjunction with human movement and settlement (Auffray et al. 1990; Guenet and Bonhomme 2003), and they have succeeded in colonizing a diverse range of environments, including alpine habitats of the Bolivian Altiplano at elevations of over 4000 m. If the ability of these mice to survive and function at such altitudes is at least partly attributable to adaptive modifications of the oxygen transport system, it seems likely that the adaptive changes would involve standing genetic variation that was carried over from the ancestral source population in Europe or Asia.
Here we report a survey of nucleotide variation in the adult globin genes of high- and low-altitude house mice from South America. Specifically, we surveyed nucleotide variation in the two closely linked α-globin paralogs on chromosome 11, HBA-T1 and HBA-T2, and the two closely linked β-globin paralogs on chromosome 7, HBB-T1 and HBB-T2. The main objectives of this study were to: (i) characterize levels and patterns of nucleotide polymorphism in the duplicated globin genes of wild house mice; (ii) test whether patterns of sequence variation are consistent with a model of long-term balancing selection; and (iii) determine whether standing variation in hemoglobin function has played a role in adaptation to recently colonized high-altitude environments in the Andes. We also collected polymorphism data from unlinked nuclear and mitochondrial loci to use as a basis for comparison with the globin genes and to shed light on the historical origins of house mice in South America.
MATERIALS AND METHODS
A total of 20 M. musculus were collected from several sea-level localities in the immediate vicinity of Lima, Peru (n = 10), and several high-altitude localities in the immediate vicinity of La Paz, Bolivia (3272–3646 m above sea level; n = 10). Genomic DNA was extracted from frozen liver tissue of each mouse using DNeasy kits (Qiagen, Valencia, CA). We surveyed nucleotide variation across the complete coding region and flanking regions of the two α-globin paralogs, HBA-T1 and HBA-T2 (both sequenced fragments were 1468 bp long), and the two β-globin paralogs, HBB-T1 and HBB-T2 (both fragments were 1477 bp long). Following the nomenclature of Aguileta et al. (2006), HBA-T1 and HBA-T2 refer to the 5′ and 3′ α-globin paralogs, respectively, and HBB-T1 and HBB-T2 refer to the 5′ and 3′ β-globin paralogs, respectively (Figure 1). Following the convention of Dickerson and Geis (1983), each amino acid residue is indexed with an alphanumerical code that designates its position relative to the eight α-helical domains (A–H) of the individual globin polypeptides.
The four globin genes were PCR-amplified using Ampli-taq Gold chemistry (Applied Biosystems, Foster City, CA) under the following cycling parameters: 94° (120 sec) initial denaturing, [94° (30 sec), 58° (30 sec), 72° (60 sec)] 30×, and a final extension at 72° (120 sec). The same PCR primers were also used for sequencing reactions, although the annealing temperature was increased to 60°. After first obtaining direct sequence data for each of the four globin genes, we then cloned both alleles from all individuals that were heterozygous at multiple sites. Specifically, we cloned diploid PCR products into pCR4-TOPO vector (Invitrogen, Carlsbad, CA) and we then sequenced six or more clones per locus. M13 primers were used to amplify cloned products (55° annealing temperature) and internal T7/T3 primers were used for sequencing. By sequencing diploid PCR products as well as cloned alleles, it is possible to control for singleton nucleotide changes that are introduced by cloning and sequencing errors. In our sequence data, all singleton nucleotide changes that were initially identified as heterozygous sites in the direct sequencing results were confirmed as true mutations if they were subsequently recovered in one or more cloned alleles. For each of the four globin genes, the exact haplotype phase of all heterozygous sites was determined experimentally. We therefore obtained complete diploid genotypes for all mice in the sample.
For the purpose of assessing the genealogical structure of the sample of house mice we used mitochondrial DNA sequence variation from the complete cytochrome b gene (1144 bp) and the control region (1063 bp, including the flanking tRNAs), yielding a concatenated alignment of 2207 bp. For the phylogenetic analysis, we added mtDNA sequences from the 20 South American specimens to an alignment that included a global reference sample of M. m. musculus, M. m. domesticus, and several closely related species of Mus. We assembled different reference panels for cytochrome b and control region sequences. All mtDNA sequences in the reference panels were downloaded from GenBank. The cytochrome b reference sequences were taken from the studies of Lundrigan et al. (2002), Pfunder et al. (2004), Suzuki et al. (2004), and Terashima et al. (2006), and the control region reference sequences were taken from the studies of Prager et al. (1993, 1998). Specimen identification numbers and GenBank accession numbers for all mice included in the reference panels are listed in supplemental Table 1 at http://www.genetics.org/supplemental/. We also included homologous sequence data from the C57BL/6J and BALB/cByJ inbred strains (Bayona-Bafaluy et al. 2003; Acin-Perez et al. 2004).
For the purpose of making comparisons with the 4 globin genes, we also surveyed variation in an additional nuclear locus, intron 7 of β-fibrinogen (718 bp), which is located at map position 48.2 cM on chromosome 3. After first obtaining direct sequence data for the β-fibrinogen intron, we then cloned both alleles from all individuals that were heterozygous at multiple sites. As with the globin genes, the haplotype phase of all heterozygous sites was determined experimentally.
All sequencing reactions and samples were run on an ABI 3730 capillary sequencer using Dye Terminator chemistry (Applied Biosystems). For each of the nuclear and mitochondrial loci, primer sequences and nucleotide positions of sequenced gene regions are given in supplemental Table 2 at http://www.genetics.org/supplemental/. All sequences have been deposited in GenBank (accession nos. EF605348–EF605508).
In addition to measuring variation within the sample of South American mice, we also estimated nucleotide divergence to orthologous sequences from the brown Norway rat, Rattus norvegicus, and two inbred strains of M. musculus, C57BL/6J and BALB/cByJ. The Rattus α-globin sequences were taken from the NCBI genome build 3.4 (accession no. NW_047334) and β-globin sequences were taken from the Ensemble genome assembly RGSC 3.4 (accession nos. ENSROG00000033465 and 29719 for HBB-T1 and HBB-T2, respectively). C57BL/6J sequences were taken from the NCBI genome build 36.1 (accession nos. NT_039515 for the α-globin genes and NT_039433 for the β-globin genes), and BALB/cByJ sequences were taken from a clone-based, alternate assembly of NCBI genome build 36.1 (accession no. NT_095534).
GENETIC DATA ANALYSIS
Alignment:
DNA sequences were aligned and contigs were assembled using ClustalW (Thompson et al. 1994). All sequences were verified by visual inspection of chromatograms. All alignments were straightforward with the exception of HBB-T2. A portion of the second intron of this gene could not be unambiguously aligned due to a high density of insertions and deletions, and the corresponding sites (positions 827–1277 in the alignment) were therefore treated as missing data.
Analysis of phylogenetic relationships:
We reconstructed mtDNA phylogenies of house mice using neighbor-joining and maximum likelihood tree-building methods. Neighbor-joining analyses were based on the matrix of uncorrected p-distances and were conducted with the program MEGA ver 3.1 (Kumar et al. 2004). The maximum-likelihood tree searches were conducted in Treefinder ver. May 2006 (Jobb et al. 2004), and this program was also used to estimate parameters of a GTR+Γ model of nucleotide substitution. In the maximum likelihood analyses, we used three independent data partitions for each codon position in the cytochrome b alignment, and we used a single partition for the control region alignment. We evaluated support for the relevant nodes with 100 bootstrap replicates (Felsenstein 1985).
Analysis of population structure:
To test for evidence of nucleotide differentiation between the high- and low-altitude samples, we conducted locus-specific permutation tests based on two summary statistics: KST* (Hudson et al. 1992) and Snn (Hudson 2000). The KST* statistic provides a measure of sequence differentiation based on the partitioning of nucleotide diversity within and between samples. The Snn statistic measures how often haplotypes that are nearest neighbors in sequence space are derived from the same sample locality. To test for differences in the frequencies of alternative protein alleles between the high- and low-altitude samples, we performed exact tests on contingency tables of amino acid haplotypes (Raymond and Rousset 1995).
Detection of recombination and gene conversion:
To detect evidence of intragenic recombination, we used the four-gamete test of Hudson and Kaplan (1985) to estimate the minimum number of recombination events in the history of the sample. To detect evidence of gene conversion between paralogous gene duplicates on the same chromosome and to estimate the number and length of ectopic conversion tracts within each locus, we used the method of Betran et al. (1997). Specifically, we aligned sequences from each pair of globin paralogs and used a maximum likelihood algorithm to estimate ψ, the per-site probability of detecting a gene conversion event between the two paralogous sequences. Conversion tract lengths were then estimated as L = TR − TL + 1 − G, where TL(5′) and TR(3′) are the site positions of the outermost informative sites within a congruent tract of nucleotide polymorphisms that are shared between the two paralogs, and G is the number of alignment gaps between TL and TR.
Analysis of nucleotide polymorphism:
We estimated two different measures of DNA sequence variation: nucleotide diversity, π (Nei and Li 1979), which is based on the average number of pairwise differences between sampled alleles, and θW (Watterson 1975), which is based on the total number of segregating sites in the sample. To assess whether observed distributions of allele frequencies deviated from the expectations of a neutral-equilibrium model, we conducted neutrality tests based on Tajima's (1989) D statistic. This statistic measures the standardized difference between π and θW, which provide independent estimates of the neutral parameter θ (= 4Nμ, where N is the effective population size and μ is the per-nucleotide mutation rate). To obtain critical values for Tajima's D, we generated a null distribution of the test statistic by conducting 10,000 coalescent simulations under a Wright-Fisher model. The simulations were conditioned on the observed number of segregating sites. We used the same simulation-based approach to perform the haplotype tests of Depaulis and Veuille (1998), which assess whether the observed haplotype number (K-test) and haplotype diversity (H-test) conform to the expectations of a neutral-equilibrium model. To measure levels of intragenic linkage disequilibrium (LD), we assessed the significance of pairwise associations between informative, biallelic nucleotide polymorphisms using Fisher's exact test with Bonferroni-adjusted critical values. To assess whether the observed levels of intragenic LD deviated from the expectations of a neutral-equilibrium model, we conducted neutrality tests based on Kelly's (1997) Zns statistic. As with the neutrality tests based on Tajima's D, haplotype number, and haplotype diversity, we obtained critical values for the Zns statistic by conducting 10,000 coalescent simulations that were conditioned on the observed number of segregating sites. The simulated genealogies were generated with no recombination, which makes the test conservative for the purpose of rejecting neutrality. For each combination of linked paralogs, we tested the null hypothesis that genotypes at one locus were independent of genotypes at the other locus by performing exact tests on contingency tables of diploid genotypes.
To assess whether ratios of polymorphism to divergence deviated from the expectations of a neutral model, we conducted a multilocus Hudson–Kreitman–Aguadé (HKA) test (Hudson et al. 1987) using the nuclear loci and the concatenated mtDNA sequences. We jointly estimated neutral parameters from all six loci to obtain expected values for the HKA test, and we then used the resultant parameter estimates to conduct coalescent simulations using the HKA program (http://lifesci.rutgers.edu/∼heylab/HeylabSoftware.htm). Orthologous sequence from Rattus was used to estimate locus-specific levels of nucleotide divergence. Summary statistics of nucleotide polymorphism, divergence, and LD were computed with the programs SITES (Hey and Wakeley 1997) and DnaSP v4.10 (Rozas et al. 2003).
RESULTS
Ancestry and population structure:
Maximum likelihood and neighbor-joining phylogenetic analyses of mtDNA sequences indicate that all specimens collected from Lima and La Paz are referable to the western European subspecies M. m. domesticus (Figure 2). Trees based on the control region and cytochrome b sequences were very similar, as were the levels of bootstrap support for corresponding nodes, although the control region tree provided higher resolution of relationships among M. m. domesticus specimens. The control region and cytochrome b trees both revealed that the South American mice fell into two separate clades with high bootstrap support. In the control region tree (Figure 2A), one clade contained all 10 Lima specimens, 6/10 La Paz specimens, and 1 reference specimen from Peru, and the other clade contained the remaining La Paz specimens. The same La Paz specimens also fell in different clades in the cytochrome b tree (Figure 2B). Average genetic distances between these clades were 1.1% for the control region tree and 0.9% for the cytochrome b tree.
Figure 2.—
Phylogenetic affinities of South American house mouse specimens (shown in bold) in relation to a global reference sample of M. musculus and congeneric species. (A) Neighbor-joining tree based on cytochrome b sequence variation (1144 bp). (B) Neighbor-joining tree based on mtDNA control region sequence variation (1063 bp). Bootstrap support values are given for select nodes on each tree.
Since house mice colonized the New World within the past several hundred years, the mtDNA phylogeny of our South American sample primarily captures the genealogical structure of the ancestral source population. The genealogical structure of our sample of haplotypes is not likely to be informative about recent historical population structure in South America because there is not a sufficient level of mutational resolution. The fact that our sample of South American mice fell into two distinct clades could indicate that these mice (or at least the mice from La Paz) represent a heterogeneous mixture of M. m. domesticus that are descendents of genetically distinct founding stocks. However, surveys of mtDNA, Y-chromosome, and X-chromosome variation in European M. musculus have revealed no relationship between geography and phylogeny, and highly divergent haplotypes are often sampled from the same locality (Tucker et al. 1989; Nachman et al. 1994; Nachman 1997). Moreover, silent site diversity in our pooled sample of mtDNA haplotypes from Lima and La Paz is very similar to values reported for broad regional areas within Europe (0.0060 vs. 0.0069; Nachman et al. 1994). Thus, the level of mtDNA variation in our sample of South American mice is similar to that observed in population samples of M. m. domesticus from the species' ancestral range.
Comparison of the high- and low-altitude samples (La Paz and Lima, respectively) revealed significant levels of nucleotide differentiation at three of the polymorphic loci: HBA-T1, β-fibrinogen, and mtDNA. By contrast, no significant differentiation was evident at HBB-T1 or HBB-T2 (Table 1).
TABLE 1.
Permutation tests of nucleotide differentiation between high- and low-altitude samples
| Locus | KST* | Snn |
|---|---|---|
| Globin genes | ||
| HBA-T1 | 0.2978 (P < 0.0001) | 0.7355 (P < 0.0001) |
| HBB-T1 | −0.0018 (P = 0.4040) | 0.5324 (P = 0.2180) |
| HBB-T2 | 0.0129 (P = 0.2260) | 0.5197 (P = 0.2550) |
| Unlinked loci | ||
| β-Fibrinogen | 0.2158 (P = 0.0090) | 0.6071 (P = 0.0080) |
| Cytochrome b + control region | 0.1792 (P < 0.0001) | 1.0000 (P < 0.0001) |
The HBA-T2 gene was invariant and was therefore not included in the analysis.
Levels and patterns of nucleotide variation at nuclear genes:
In the total sample of 20 mice, the average level of silent site diversity at the 5 nuclear loci was π = 0.0063 and ranged from a low of π = 0.0000 at HBA-T2 to a high of π = 0.0180 at HBB-T2. Average silent site diversity based on the number of segregating sites was θ = 0.0069 and ranged from a low of θ = 0.0000 at HBA-T2 to a high of θ = 0.0199 at HBB-T1. The two β-globin paralogs, HBB-T1 and HBB-T2, were characterized by much higher levels of nucleotide polymorphism than the remaining three nuclear loci (Table 2).
TABLE 2.
Summary of nucleotide polymorphism at nuclear and mitochondrial loci in South American house mice
| Length (bp) | Sample | Na | Sb | Hc | HDd | π(silent) | θW(silent) | K(silent)e | π/K | |
|---|---|---|---|---|---|---|---|---|---|---|
| Globin genes | ||||||||||
| HBA-T1 | 1468 | Lima | 20 | 4 | 3 | 0.542 | 0.0011 | 0.0006 | ||
| La Paz | 20 | 3 | 4 | 0.553 | 0.0007 | 0.0009 | ||||
| Total | 40 | 7 | 6 | 0.663 | 0.0013 | 0.0013 | 0.2021 | 0.0065 | ||
| HBA-T2 | 1468 | Lima | 18 | 0 | 1 | 0.000 | 0.0000 | 0.0000 | ||
| La Paz | 20 | 0 | 1 | 0.000 | 0.0000 | 0.0000 | ||||
| Total | 38 | 0 | 1 | 0.000 | 0.0000 | 0.0000 | 0.2764 | — | ||
| HBB-T1 | 1477 | Lima | 16 | 62 | 15 | 0.992 | 0.0106 | 0.0148 | ||
| La Paz | 20 | 63 | 17 | 0.979 | 0.0096 | 0.0130 | ||||
| Total | 36 | 107 | 31 | 0.989 | 0.0099 | 0.0199 | 0.1538 | 0.0644 | ||
| HBB-T2 | 1477 | Lima | 18 | 38 | 6 | 0.810 | 0.0183 | 0.0112 | ||
| La Paz | 18 | 44 | 9 | 0.804 | 0.0169 | 0.0134 | ||||
| Total | 36 | 47 | 11 | 0.810 | 0.0180 | 0.0122 | 0.1671 | 0.1126 | ||
| Unlinked loci | ||||||||||
| β-fibrinogen, intron 7 | 718 | Lima | 14 | 3 | 3 | 0.473 | 0.0016 | 0.0013 | ||
| La Paz | 14 | 3 | 3 | 0.473 | 0.0017 | 0.0013 | ||||
| Total | 28 | 3 | 4 | 0.587 | 0.0022 | 0.0011 | 0.2035 | 0.0106 | ||
| Cyt b + control region | 2207 | Lima | 10 | 5 | 5 | 0.822 | 0.0019 | 0.0016 | ||
| La Paz | 10 | 22 | 4 | 0.778 | 0.0077 | 0.0053 | ||||
| Total | 20 | 27 | 9 | 0.905 | 0.0060 | 0.0057 | 0.3085 | 0.0196 |
Number of sampled chromosomes.
Number of segregating sites.
Number of haplotypes.
Haplotype diversity.
Jukes-Cantor corrected divergence to Rattus.
Observed levels of silent site diversity at HBA-T1, HBA-T2, and β-fibrinogen (mean π = 0.0012, range = 0.0000–0.0022) were very similar to levels of variation that have been reported for noncoding autosomal and X-linked loci in samples of M. m. domesticus from western Europe and southwestern Asia (Nachman 1997; Harr 2006; Baines and Harr 2007) (Table 3). This pattern is consistent with the results of allozyme surveys that indicate that levels of heterozygosity in New World house mice are typically commensurate with levels observed in samples of M. m. domesticus from western Europe (Selander and Yang 1969; Selander et al. 1969a,b; Sage 1981).
TABLE 3.
Levels of nucleotide diversity at noncoding nuclear loci in population samples of Old World M. musculus domesticus
| Source | Chromosome | No. of loci | Mean no. bp/locus | Mean no. sampled chromosomes/locus | Mean π (range) | Reference |
|---|---|---|---|---|---|---|
| Italy | X linked | 4 | 1505.5 | 10.0 | 0.0010 (0.0000–0.0021)a | Nachman (1997) |
| Germany | X linked | 6 | 522.5 | 7.8 | 0.0007 (0.0000–0.0009)a | Baines and Harr (2007) |
| France | X linked | 6 | 522.5 | 7.8 | 0.0001 (0.0000–0.0008)a | Baines and Harr (2007) |
| Iran | X linked | 6 | 522.0 | 11.0 | 0.0016 (0.0000–0.0032)a | Baines and Harr (2007) |
| Germany | Autosomal | 14 | 595.7 | 15.4 | 0.0025 (0.0000–0.0073) | Harr (2006) |
| Germany | Autosomal | 7 | 959.3 | 15.7 | 0.0013 (0.0000–0.0033) | Baines and Harr (2007) |
| France | Autosomal | 7 | 955.3 | 16.0 | 0.0026 (0.0002–0.0074) | Baines and Harr (2007) |
| Iran | Autosomal | 7 | 1080.4 | 15.7 | 0.0033 (0.0007–0.0059) | Baines and Harr (2007) |
Reported values were multiplied by 4/3 to account for different effective population sizes of X-linked and autosomal loci.
Although levels of silent site diversity in mtDNA and three nuclear genes (HBA-T1, HBA-T2, and β-fibrinogen) were closely similar to reported estimates from Old World M. m. domesticus (Nachman et al. 1994; Nachman 1997; Harr 2006; Baines and Harr 2007), levels of silent site diversity at the two β-globin paralogs, HBB-T1 and HBB-T2, were surprisingly high. Observed silent site diversities at HBB-T1 and HBB-T2 in the sample of South American mice (π = 0.0099 and 0.0180, respectively) exceeded the upper range of values reported for population surveys of nuclear genes in Old World M. m. domesticus (Table 3) and also exceeded the upper range of silent site diversities for 19 nuclear loci that were surveyed in a pooled sample of wild-derived inbred strains of M. musculus that were representative of different subspecies (mean π = 0.0041, range = 0.0000–0.0081; Takahashi et al. 2004). Results of a multilocus HKA test revealed that the observed disparity in silent site diversity between HBB-T1 and the other four unlinked loci cannot be reconciled under a neutral model of molecular evolution (Table 4). Although HBB-T2 exhibited a similar deviation from neutral expectations, this locus violates assumptions of the HKA test because most of the segregating variation at HBB-T2 is attributable to gene conversion from the paralogous HBB-T1 gene (discussed below). For this reason, we have excluded HBB-T2 from analyses that are based on the infinite-sites mutation model.
TABLE 4.
Multilocus HKA test involving all loci other than HBB-T2
| Polymorphism
|
Divergence
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Locus | I | θ (2.5–97.5%) | S/ES | Var | Dev | D/ED | Var | Dev |
| HBB-T1 | 1.00 | 0.0053 (0.0036–0.0076) | 81/30.65 | 118.98 | 21.31 | 141/191.35 | 244.66 | 10.36 |
| HBA-T1 | 1.00 | 0.0042 (0.0029–0.0060) | 5/25.30 | 82.58 | 4.99 | 156/135.70 | 162.51 | 2.54 |
| HBA-T2 | 1.00 | 0.0054 (0.0036–0.0077) | 0/31.79 | 124.44 | 8.12 | 203/171.21 | 213.89 | 4.73 |
| β-fibr | 1.00 | 0.0053 (0.0034–0.0083) | 3/14.84 | 38.22 | 3.67 | 54/42.16 | 44.75 | 3.31 |
| mtDNA | 0.25 | 0.0062 (0.0042–0.0088) | 25/11.43 | 27.96 | 6.59 | 300/313.57 | 323.06 | 0.57 |
 P < 0.000001. I, inheritance scalar (= 1.00 for autosomal loci, and 0.25 for mtDNA); S, observed number of segregating sites; ES, expected number of segregating sites; D, observed divergence to Rattus; ED, expected divergence.
P < 0.000001. I, inheritance scalar (= 1.00 for autosomal loci, and 0.25 for mtDNA); S, observed number of segregating sites; ES, expected number of segregating sites; D, observed divergence to Rattus; ED, expected divergence.
In addition to the higher-than-expected level of silent site diversity at HBB-T1, this gene was also characterized by a significant excess of low-frequency polymorphisms (Tajima's D = −1.893, P = 0.0120). By contrast, the other unlinked polymorphic loci were characterized by an excess of intermediate-frequency polymorphisms: Tajima's D values ranged from 0.277 for the concatenated mtDNA sequence to 2.435 for β-fibrinogen. In fact, the observed excess of intermediate-frequency polymorphisms at β-fibrinogen was statistically significant (P = 0.0020). Thus, the distributions of allele frequencies at HBB-T1 and the other unlinked loci were skewed in completely opposite directions. Only 0.57% of the simulated data sets from the multilocus HKA test exhibited a higher variance of Tajima's D values. The HBB-T1 gene was also characterized by an excess number of haplotypes (K-test, P = 0.0050) and a nearly significant excess of haplotype diversity (H-test, P = 0.0690), a pattern that was not observed at the other unlinked loci. The fact that HBB-T1 and the other unlinked loci are characterized by such different site-frequency distributions and haplotype distributions is consistent with results of the multilocus HKA test in suggesting that the observed departures from neutral-equilibrium expectations are attributable to effects that are specific to HBB-T1.
Amino acid variation, haplotype structure, and gene conversion:
Two distinct α-globin protein haplotypes were observed in the sample of South American mice. There was a single fixed amino acid difference between the HBA-T1 and HBA-T2 paralogs, 68(E17)Ser→Asn, and there were two amino acid polymorphisms in HBA-T1, 25(B6)Gly/Val, and 62(E11)Val/Ile, that were monomorphic in HBA-T2 (Figure 3). Consequently, these mice are capable of synthesizing three distinct α-chain polypeptides, two of which are encoded by HBA-T1 and one that is encoded by the monomorphic HBA-T2 paralog. Each of these amino acid sequences has been previously described in different inbred strains of M. musculus. The two-locus α-globin haplotype containing the ancestral ‘25Gly/62Val’ allele at HBA-T1 is fixed in the SM/J strain, and the alternative haplotype containing the derived ‘25Val/62Ile’ allele at HBA-T1 is fixed in the SWR/J strain (Hilse and Popp 1968; Popp et al. 1982; Erhart et al. 1987). According to the conventional nomenclature of mouse genetics, the latter two-locus haplotype is referred to as the Hbad haplotype and the former is referred to as the Hbac haplotype (Erhart et al. 1987). There was no evidence of recent gene conversion between the two α-globin paralogs of the South American mice, as none of the nucleotide polymorphisms in HBA-T1 were segregating a derived variant that was characteristic of the HBA-T2 haplotype background. In the sample of HBA-T1 alleles there was no evidence of intragenic recombination or interallelic gene conversion. Consistent with the apparently low rate of recombination at HBA-T1, significant LD was detected at 6 of 15 pairwise comparisons between informative nucleotide polymorphisms. However, the overall level of intragenic LD did not exceed neutral-equilibrium expectations (Zns = 0.3977, P > 0.05).
Figure 3.—
Structural alignment of mammalian α- and β-globin sequences. For M. musculus, the alignment includes representative c- and d-type protein alleles from the two α-globin paralogs and representative s- and d-type protein alleles from the two β-globin paralogs. For the β-globin sequences, redox-active cysteine residues are shown in boxes.
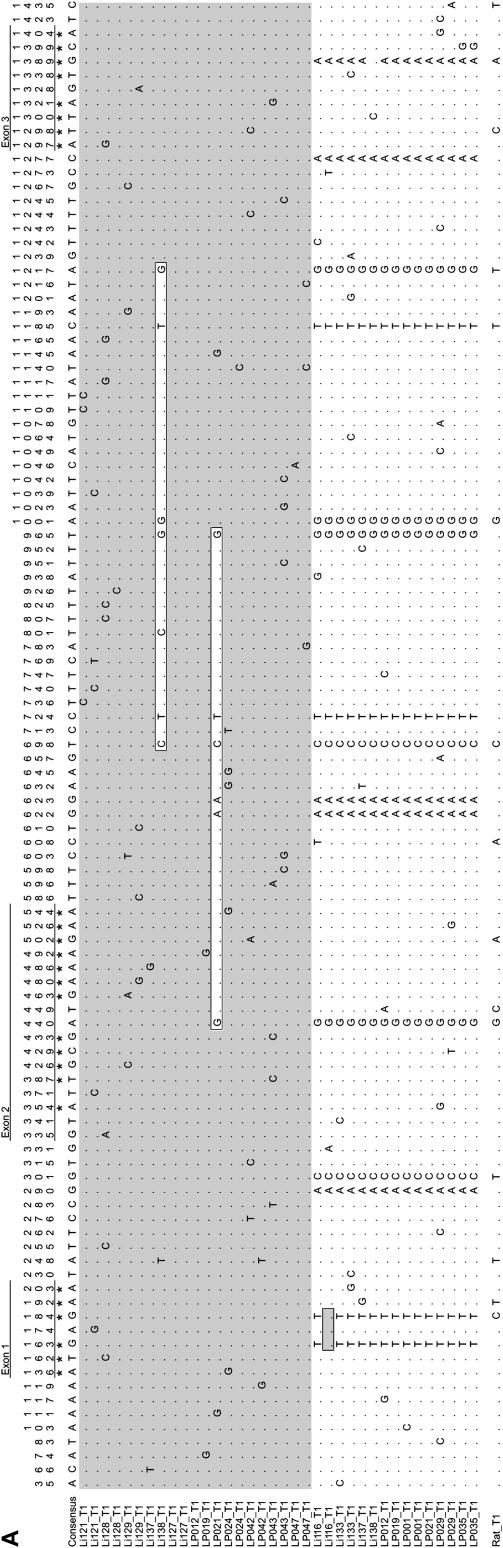
Similar to the case with the α-globin paralogs, two main β-globin protein haplotypes were observed in the sample of South American mice. The allelic and nonallelic patterns of β-globin amino acid variation are graphically summarized in Figure 4. One of the inferred haplotypes contained two β-globin paralogs that encode identical polypeptides, while the other haplotype contained two paralogs that were distinguished by nine amino acid substitutions: 9(A6)Ala → Ser, 16(A13)Gly → Ala, 20(B2)Ser → Ala/Pro, 58(E2)Ala → Pro, 73(E17)Asp →Glu, 76(EF1)Asn → Lys, 77(EF2)His → Asn, 80(EF5)Ser →Asn, and 109(G11)Met → Ala (Figure 3). Consequently, these mice are capable of synthesizing three distinct β-globin polypeptides, one of which is encoded by identical HBB-T1 and HBB-T2 paralogs on the same haplotype, and the remaining two are encoded by distinct paralogs on the alternative haplotype. Each of these two-locus β-globin haplotypes has been previously described in different inbred strains of M. musculus. The two-locus haplotype with identical HBB-T1 and HBB-T2 paralogs (the Hbbs haplotype) is fixed in the C57BL/10 strain and the alternative two-locus haplotype with the distinct HBB-T1 and HBB-T2 paralogs (the Hbbd haplotype) is fixed in the BALB/cByJ strain (Erhart et al. 1985). These two-locus β-globin haplotypes also encode the HBB-D and HBB-S protein electromorphs that have been studied extensively in populations of M. musculus throughout the world (Petras 1967; Selander and Yang 1969; Selander et al. 1969a,b; Berry and Murphy 1970; Myers 1974; Berry and Jakobson 1975; Berry and Peters 1975, 1977, 1981; Berry et al. 1978; Sage 1981; Sage et al. 1986). At the HBB-T1 gene, the s- and d-type alleles are distinguished by three nearly fixed amino acid differences: 13(A10)Gly → Cys, 20(B2)Ser → Ala, and 139(H17)Thr → Ala (Figure 4). These amino acid substitutions are due to nonsynonymous changes at positions 163, 184, and 1389 (Figure 5A) and are henceforth referred to as fixed differences, although in each case the frequency of the minor SNP allele is 1/15 in the d-type allele class. At the HBB-T2 gene, the s- and d-type alleles are distinguished by 11 fixed amino acid differences: 9(A6)Ala → Ser, 13(A10)Gly → Cys, 16(A13)Gly → Ala, 20(B2)Ala → Pro, 58(E2)Ala → Pro, 73(E17)Asp → Glu, 76(EF1)Asn → Lys, 77(EF2)His → Asn, 80(EF5)Ser →Asn, 109(G11)Met → Ala, and 139(H17)Thr → Ala (Figure 4). These fixed differences between s- and d-type alleles at both β-globin paralogs represent the best candidate sites for selection if alternative protein alleles are in fact maintained as a balanced polymorphism.
Figure 4.—
Schematic illustration of allelic and nonallelic variation in the two house mouse β-globin paralogs, HBB-T1 and HBB-T2, on each of two haplotype backgrounds, Hbbs and Hbbd. Numbers refer to pairwise amino acid differences between alleles or paralogous genes. In the sample of South American mice, HBB-T1 and HBB-T2 are in nearly complete LD with each other such that chromosomes that carry a d-type allele at HBB-T1 typically carry a d-type allele at HBB-T2 (the Hbbd haplotype), and likewise for s-type alleles (the Hbbs haplotype).
Figure 5.—
Nucleotide polymorphism in the two β-globin paralogs of South American house mice, HBB-T1 and HBB-T2. For both genes, s-type alleles are shown in gray and d-type alleles are shown in white. Nonsynonymous polymorphisms are denoted by asterisks at the top of the alignments. In the HBB-T1 table of polymorphic sites (A), three interallelic gene conversion tracts are shown in boxes. In the HBB-T2 table of polymorphic sites (B), a dagger denotes the position of the localized cluster of insertion/deletion differences between the s- and d-type alleles (nucleotide positions 827–1277) that results in unalignable sequence between the two allele classes.
The haplotype dichotomy observed at the amino acid level is also clearly evident at the nucleotide level (Figure 5), and the distinction between s- and d-type alleles at both β-globin paralogs is readily apparent in distance-based phylogenetic trees (Figure 6). Comparisons between s- and d-type allele classes revealed that the sample of s-type alleles at HBB-T1 was characterized by a higher level of nucleotide polymorphism and lower LD (Table 5), suggesting that the s-type allele may have been present at a higher relative frequency in the past. The sample of d-type alleles at HBB-T2 was the only allele class that did not exhibit a significant excess of low-frequency polymorphisms (Table 5), which likely reflects the different evolutionary histories of converted and nonconverted alleles.
Figure 6.—
Neighbor-joining phylogenies of s- and d-type alleles from each of the two β-globin paralogs, HBB-T1 (A) and HBB-T2 (B). In A, the three most basal branches in the clade of d-type alleles (specimen IDs shown in bold with an asterisk) represent sequences that have undergone interallelic gene conversion. ‘La Paz 021 HBB-T1 A’ and ‘Lima 138 HBB-T1 B’ are s-type alleles that have been partially converted by a d-type sequence, whereas ‘Lima 116 HBB-T1 A’ is a d-type allele that has been partially converted by an s-type sequence. See Figure 5A for details regarding the length and composition of the conversion tracts.
TABLE 5.
Levels and patterns of intra-allelic nucleotide polymorphism and LD at house mouse β-globin paralogs
| Locus | Allele class | N | S | πTotal | πSilent | θTotal | θSilent | Tajima's D | Zns |
|---|---|---|---|---|---|---|---|---|---|
| HBB-T1 | s | 21 | 68 | 0.0050 | 0.0055 | 0.0138 | 0.0150 | −2.560* | 0.062 |
| d | 15 | 37 | 0.0036 | 0.0033 | 0.0082 | 0.0075 | −2.367* | 0.112 | |
| HBB-T2 | s | 16 | 5 | 0.0014 | 0.0020 | 0.0015 | 0.0023 | −0.379 | 0.126 |
| d | 20 | 10 | 0.0011 | 0.0012 | 0.0028 | 0.0033 | −2.145* | 0.114 |
Estimates of Zns are based on the total number of segregating sites within each allele class. *P < 0.05.
Results of the four-gamete test revealed evidence for a single recombination event in the history of sampled HBB-T1 alleles and zero recombination events in the history of HBB-T2 alleles. Consistent with these apparently low rates of intragenic recombination, 105 of 153 pairwise comparisons between informative nucleotide polymorphisms exhibited significant LD at HBB-T1 and 528 of 703 such comparisons were significant at HBB-T2. Each of the two β-globin paralogs exhibited a highly significant excess of intragenic LD (HBB-T1: Zns = 0.601, P = 0.0100; HBB-T2: Zns = 0.774, P = 0.0010). In the case of HBB-T2, the excess LD is attributable to nonrandom associations between converted and nonconverted alleles (see below). In addition to the high levels of intragenic LD, HBB-T1 and HBB-T2 were also characterized by a highly significant level of intergenic LD with respect to s- and d-type alleles (P = 0.0002, Fisher's exact test). Consequently, the majority of chromosomes that carry a d-type allele at one paralog also carry a d-type allele at the other paralog, and likewise for s-type alleles. For 17 of the mice in our sample, we recovered complete two-locus genotypes for both β-globin paralogs, and in 3 cases a mouse that was heterozygous for s-type and d-type alleles at one paralog was homozygous at the other paralog. The remaining 14 mice were either double heterozygotes or double homozygotes, suggesting that recombinant Hbbd/Hbbs haplotypes are present at a frequency of <10%.
Haplotype-specific differences in divergence between HBB-T1 and HBB-T2 are clearly attributable to haplotype-specific differences in the history of interparalog gene conversion. As shown in Figure 7A, a phylogenetic reconstruction based on an alignment of flanking sequence that spans the 5′ UTR (nucleotides 1–123) accurately recovers the true orthologous relationships between the β-globin genes of Mus and Rattus. By contrast, a phylogenetic reconstruction based on an alignment of intron 2 (nucleotides 1130–1280) groups the s-type allele of HBB-T2 with paralogous HBB-T1 sequences (Figure 7B). On the Hbbs haplotype, gene conversion has completely homogenized sequence variation between HBB-T1 and HBB-T2, and the level of interparalog nucleotide divergence across the coding region is uniformly low. As can be seen in Figure 8A, levels of nucleotide divergence increase dramatically in the 5′ and 3′ flanking regions of the coding sequence. The abrupt increases in sequence divergence demarcate the 5′ and 3′ boundaries of the gene conversion tract, which are located just outside of the translational start and stop codons (Erhart et al. 1987). On the Hbbd haplotype, by contrast, there is no evidence of gene conversion between HBB-T1 and HBB-T2 and the two paralogs are much more divergent (Figure 8B). Due to the different histories of gene conversion on the two haplotype backgrounds, average nucleotide divergence between the two paralogs on the Hbbd background is nearly an order of magnitude higher than on the Hbbs background (Dxy = 0.0400 vs. 0.0053). As pointed out by Erhart et al. (1987), concerted evolution of HBB-T1 and HBB-T2 on the Hbbs haplotype produces a highly unusual situation in which the two paralogs are more similar to one another than either one is to its allelic counterpart on the Hbbd haplotype (Figure 4). Although we cannot definitively rule out the possibility that HBB-T1 has been on the receiving end of interparalog gene conversion at some point in its evolutionary history, the only conversion events that we detected involved HBB-T1 as the donor and HBB-T2 as the recipient. In addition to the high level of sequence similarity between HBB-T1 and HBB-T2 on the Hbbs haplotype, some of the sequence similarity between the HBB-T1 allele on the Hbbd haplotype and the HBB-T1 allele on the Hbbs haplotype is attributable to interallelic gene conversion. The algorithm of Betran et al. (1997) identified two HBB-T1 s-type alleles that harbored conversion tracts that were introduced from the d-type background and one HBB-T1 d-type allele that appears to have been partially converted by an s-type allele (Figure 5A). These interallelic conversion events presumably involved recombination between nonsister chromatids in Hbbs/Hbbd heterozygotes.
Figure 7.—
Alternative neighbor-joining phylogenies that illustrate the effects of concerted evolution between HBB-T1 and HBB-T2 on the Hbbs haplotype. In A, the phylogeny based on 5′ flanking sequence recovers orthologous relationships among rodent HBB-T1 and HBB-T2 sequences. In B, by contrast, the phylogeny based on intronic sequence does not recover the true orthologous relationships due to conversion of HBB-T2 by HBB-T1 on the Hbbs haplotype.
Figure 8.—
Sliding window plots of average nucleotide divergence between the two β-globin paralogs on the Hbbs haplotype (A) and the Hbbd haplotype (B). Window size, 50 bp; step size, 10 bp.
With the exception of the monomorphic HBA-T2 gene, each of the other three globin genes was characterized by a surprisingly high level of replacement polymorphism relative to synonymous polymorphism (Table 6). If alternative protein alleles are maintained as a balanced polymorphism at HBA-T1, HBB-T1, or HBB-T2, then this might be reflected by an elevated ratio of replacement:synonymous polymorphism relative to the ratio of replacement:synonymous fixed differences between species (Mcdonald and Kreitman 1991). Although McDonald–Kreitman tests were not significant for HBA-T1 or HBB-T2, the test revealed a significant excess of replacement polymorphism at HBB-T1 (P = 0.046, Fisher's exact test). However, only 3 of the 23 replacement polymorphisms at HBB-T1 were present at frequencies >10% because they represent fixed differences between the intermediate-frequency s- and d-type alleles: 13(A10)Gly/Cys, 20(B2)Ala/Ser, and 139(H17)Ala/Thr (Figure 5A).
TABLE 6.
Polymorphism and divergence in the duplicated globin genes of South American house mice
| No. of sites
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Coding
|
Polymorphism within M. musculus
|
Divergence to Rattusa
|
|||||||||
| Total lengthb | N | Nonsynonymous | Synonymous | S | πA | πS | πA/πS | KA | KS | KA/KS | |
| HBA-T1 | 1468 (1225) | 40 | 320.97 | 105.03 | 7 | 0.0027 | 0.0009 | 2.961 | 0.0897 | 0.2397 | 0.374 |
| HBA-T2 | 1468 (1220) | 38 | 320.19 | 105.81 | 0 | 0.0000 | 0.0000 | – | 0.0859 | 0.2178 | 0.395 |
| HBB-T1 | 1477 (1374) | 36 | 333.66 | 107.34 | 107 | 0.0079 | 0.0095 | 0.839 | 0.0513 | 0.2102 | 0.244 |
| HBB-T2 | 1477 (984) | 36 | 332.93 | 108.07 | 47 | 0.0190 | 0.0298 | 0.639 | 0.0768 | 0.2132 | 0.360 |
Divergence estimates are Jukes-Cantor-corrected.
Total number of sites excluding alignment gaps is given in parentheses.
Levels and patterns of allelic divergence:
If the s- and d-type β-globin alleles are maintained as a long-term balanced polymorphism, population genetic theory suggests that comparisons of sequence variation between the two allele classes may reveal a pronounced peak in silent-site divergence that is centered on the selected site(s). This elevated level of divergence should then rapidly drop away as a function of recombinational distance from the site (or set of closely-linked sites) that constitutes the target of selection (Strobeck 1983; Hudson and Kaplan 1988; Kelly and Wade 2000; Slatkin 2000; Navarro and Barton 2002; Nordborg and Innan 2003; Charlesworth 2006). In the case of HBB-T1, 2 of the fixed amino acid differences between the s- and d-type alleles are associated with a peak in silent site divergence in exon 1 (Figure 9). However, this peak is one of several equal-sized peaks that are spaced along the gene, so patterns of nucleotide variation provide no basis for attaching any special weight to residue positions 13 and 20 as candidate sites for balancing selection. Moreover, a sharply defined peak of divergence may not be very likely in this particular case due to the apparently low historical rate of recombination across the HBB-T1 gene region. In the case of HBB-T2, the s-type allele originated by ectopic gene conversion from the paralogous HBB-T1 gene, and in addition to the 11 fixed amino acid differences between the s- and d-type alleles, the most obvious difference between the 2 allele classes is the localized cluster of indels in intron 2 of the d-allele that renders the sequences locally unalignable.
Figure 9.—
Sliding window plot showing variation in levels of silent site diversity within and between s- and d-type alleles of the HBB-T1 gene (window size = 50 bp, step size = 10 bp). Fixed replacement differences that distinguish the two allele classes are indicated by red arrows, and fixed differences at synonymous or noncoding sites are indicated by blue arrows. The three exons of HBB-T1 are shown as gray boxes.
Altitudinal variation in house mouse hemoglobins:
In our sample of South American mice, HBA-T1 was segregating two amino acid replacement polymorphisms: 25(Gly/Val) and 62(Asn/Ser). These 2 sites were monomorphic for the ancestral variant in the high-altitude La Paz sample, but the derived variants, 25(Val) and 62(Ile), were present at frequencies of 0.65 and 0.60, respectively, in the low-altitude Lima sample. Consequently, the alternative protein alleles exhibit highly significant frequency differences between the high- and low-altitude samples (P < 0.0001, Fisher's exact test). The two amino acid sites were in nearly complete LD in the Lima sample (D' = 1.00, P < 0.0001), reflecting the fact that the derived residues were in coupling phase in all but one of the three observed gametic types. As first pointed out by Erhart et al. (1987), the α-globin haplotype defined by the derived 25(B6)Val and 62(E11)Ile variants is unusual because the B6 and E11 residues are highly conserved in both hemoglobin chains and in myoglobin (Dickerson and Geis 1983). In almost all vertebrate globin chains studied to date, the B6 and E11 residues are glycine and valine, respectively. The pattern observed in Mus suggests that the paired substitutions may be compensatory (Erhart et al. 1987).
In contrast to the pattern of amino acid variation observed at HBA-T1, neither of the two β-globin paralogs exhibited any strikingly nonrandom pattern of amino acid variation with respect to altitude. In comparisons between the Lima and La Paz samples, there were no significant differences in the frequencies of s- and d-type alleles at HBB-T1 or HBB-T2 (P = 0.7431 and 0.5701, respectively; Fisher's exact test).
DISCUSSION
In the sample of South American house mice, levels of silent site diversity in mtDNA and three nuclear genes (HBA-T1, HBA-T2, and β-fibrinogen) were closely similar to reported estimates from Old World population samples of M. m. domesticus (Nachman et al. 1994; Nachman 1997; Harr 2006; Baines and Harr 2007). However, the two β-globin paralogs, HBB-T1 and HBB-T2, were characterized by levels of silent site diversity that were over an order of magnitude higher than those observed at the other unlinked nuclear genes. The surprisingly high levels of nucleotide polymorphism at HBB-T1 and HBB-T2 were attributable to the segregation of two highly divergent haplotypes, Hbbd and Hbbs. These two-locus haplotypes were present at roughly equal frequency in the total sample of South American mice and exhibited no significant frequency differences with respect to altitude.
What is the explanation for the remarkably high levels of nucleotide polymorphism at the two β-globin paralogs? In the case of HBB-T2, the elevated level of polymorphism is attributable to the fact that one of the divergent alleles is the product of wholesale gene conversion by the 5′ paralog, HBB-T1. The converted, s-type allele and the ancestral, d-type allele are present at roughly equal frequencies, and therefore most of the nucleotide variation at HBB-T2 is partitioned between the two alternative haplotype backgrounds. Since most of the segregating variation at HBB-T2 originated by a process of ectopic gene conversion, this gene violates central assumptions of the infinite-sites mutation model and therefore invalidates the results of conventional neutrality tests such as the HKA test (Innan 2003). Even though the HBB-T2 gene is not well suited to the application of neutrality tests that are based on specific mutation models, the occurrence of two highly divergent protein alleles at roughly equal frequencies is highly improbable under neutrality. A multilocus HKA test including all loci other than HBB-T2 was highly significant and showed that the most pronounced deviations from expected values were attributable to HBB-T1 (Table 4). This result suggests that the observed level of silent site polymorphism at HBB-T1 may reflect a history of long-term balancing selection.
The observed level of nucleotide divergence between s- and d-type β-globin alleles suggests that they are quite old. Since alleles that have been maintained for long periods of time will have had more opportunities to recombine, neutral theory predicts that highly polymorphic genes should also be characterized by low levels of LD (Kimura and Ohta 1973; Sabeti et al. 2002; Toomajian et al. 2003). The observed pattern of variation at the two β-globin paralogs does not conform to this neutral expectation, as the unusually high level of polymorphism is also associated with extremely high LD. This combination of high polymorphism and high LD has two possible explanations: (1) the divergent haplotypes are maintained as an ancient balanced polymorphism, or (2) the divergent haplotypes are attributable to admixture that occurred recently enough that there has not been sufficient time for LD to decay. According to the balancing selection hypothesis, the high level of polymorphism at HBB-T1 should be restricted to a relatively small chromosomal region. Even if epistatic selection is maintaining two-locus HBB-T1/HBB-T2 haplotypes as a multisite balanced polymorphism, theory predicts that the elevated polymorphism should fall away rapidly in the flanking regions upstream of the 5′ paralog (HBB-T1) and downstream of the 3′ paralog (HBB-T2) (Kelly and Wade 2000; Navarro and Barton 2002; Wiuf et al. 2004). Given that the 2 paralogs are separated by 12.6 kb (Figure 1), theory also suggests that epistatic selection would have to be extremely strong to counteract the randomizing effects of recombination across such a large chromosomal region (Kelly and Wade 2000). In contrast to the balancing selection hypothesis, the recent admixture hypothesis predicts that the haplotype dichotomy observed at the β-globin paralogs should also be evident in other regions of the genome. Moreover, the divergent haplotypes could potentially extend over long physical distances.
Results of previously published studies of electrophoretic protein variation suggest that the pattern of β-globin polymorphism that we observe in the South American mice is not anomalous, as the HBB-S and HBB-D electromorphs are present at surprisingly uniform frequencies in samples of M. musculus from across the globe (Petras 1967; Selander and Yang 1969; Selander et al. 1969a,b; Berry and Murphy 1970; Wheeler and Selander 1972; Myers 1974; Berry and Jakobson 1975; Berry and Peters 1975, 1977, 1981; Berry et al. 1978; Sage 1981; Sage et al. 1986). This suggests that the unusually high levels of nucleotide polymorphism and LD that we observe at HBB-T1 and HBB-T2 are not attributable to an idiosyncratic history of introgressive hybridization that is unique to South American house mice. Instead, it is possible that the two-locus β-globin polymorphism is being maintained in South American house mice by the same form of balancing selection that is maintaining the polymorphism across the rest of the species' range.
Although the amino acid changes that distinguish the s- and d-type alleles are present at intermediate frequency, the HBB-T1 gene is otherwise characterized by an excess of low-frequency polymorphism. This pattern is paralleled at silent sites and is reflected by a significantly negative Tajima's D value within each allele class and within the pooled sample of s- and d-type alleles. Although this type of skew in the distribution of allele frequencies is not expected under a simple model of overdominant selection, negative Tajima's D values are consistent with certain models of temporally varying selection (Gillespie 1994). The fact that the s- and d-type allele classes were both characterized by an excess of low-frequency polymorphism suggests the possibility that both alleles have experienced a history of dramatic frequency changes, possibly due to some form of episodic or fluctuating selection. Temporal fluctuations in the effective sizes of the two allele classes may also explain the higher-than-expected haplotype number and haplotype diversity at HBB-T1. It appears that the β-globin genes of house mice have experienced an extremely complex history of selection and gene conversion that has had the net effect of maintaining extremely high levels of nucleotide polymorphism.
Age of the HBB polymorphism:
The remarkably high levels of nucleotide polymorphism at the two β-globin paralogs suggests that the coalescence time of Hbbs and Hbbd haplotypes may vastly exceed the coalescence times of other unlinked nuclear genes. The antiquity of the Hbbs and Hbbd haplotypes is also indicated by the fact that the β-globin polymorphism predates the divergence of the subspecies M. m. domesticus and M. m. musculus (Selander et al. 1969a; Sage et al. 1986), which are thought to have diverged ∼350,000–500,000 years ago (She et al. 1990; Suzuki et al. 2004). To determine the approximate age of the β-globin polymorphism, we estimated the time of divergence between the s- and d-type alleles of HBB-T1 by using a local molecular clock approach (Goodman 1986). As a calibration point, we used a range of Mus-Rattus divergence times spanning 15–20 MYA (Jacobs and Pilbeam 1980; Springer et al. 2003). To root the tree of Mus and Rattus HBB-T1 sequences, we used the HBB-T1 ortholog in guinea pig, Cavia porcellus, which we obtained from an unannotated genomic contig (GenBank accession no. AC181986_53698). As shown in Table 7, molecular clock estimates based on all 3 codon positions suggest that the s- and d-type alleles of M. musculus HBB-T1 are on the order of 1–2 MY old, and estimates based on 1st and 2nd codon positions suggest that the alleles are on the order of 3–4 MY old. These estimates are consistent with previous age estimates of 3–8 MY reported by Erhart et al. (1985).
TABLE 7.
Estimates of divergence time between s- and d-type HBB-T1 alleles based on a local molecular clock
| Divergence time (MYA)
|
||
|---|---|---|
| Calibration point (Mus–Rattus divergence) | All codon positions | Codon positions 1 and 2 |
| 15 MYA | Min = 0.814 | Min = 2.340 |
| Max = 1.596 | Max = 3.838 | |
| Mean = 1.216 | Mean = 3.136 | |
| 20 MYA | Min = 1.085 | Min = 3.120 |
| Max = 2.128 | Max = 5.117 | |
| Mean = 1.621 | Mean = 4.181 | |
The apparent antiquity of the β-globin haplotypes suggests that HBB-T1 and HBB-T2 would be outliers in a genome-wide distribution of coalescence times. However, it will be necessary to collect polymorphism data from additional unlinked autosomal loci to determine whether this is in fact the case. Surveys of nucleotide polymorphism among wild-derived inbred strains of house mice have revealed a surprisingly large number of ancestral polymorphisms that are shared among M. musculus subspecies (Ideraabdullah et al. 2004; Harr 2006). Of 569 biallelic SNPs and indel polymorphisms found among classical and wild-derived inbred strains of M. musculus, 37.8% of the variants arose prior to the divergence of different M. musculus subspecies (Ideraabdullah et al. 2004). These relatively ancient polymorphisms appear to be randomly distributed across the genome, and they represent 44.7% of sequence variants observed in pairwise comparisons between different M. musculus strains. Retention of ancestral polymorphism, in combination with introgressive hybridization, may account for the mosaic pattern of genomic variation in some wild-derived inbred strains of M. m. domesticus. For example, Ideraabdullah et al. (2004) noted that although strain CALB/RkJ has been assigned to the M. m. domesticus subspecies, CALB/RkJ mice possess long-range haplotypes characteristic of M. m. castaneus on several different chromosomes.
Altitudinal patterns of amino acid variation:
In our sample of mice, amino acid variation in the HBA-T1 gene was only present in the low-altitude sample from Lima and the ancestral protein allele was fixed in the high-altitude La Paz sample. Thus, even if the amino acid variation has important effects on hemoglobin function, the geographic pattern of variation in our sample does not suggest any obvious role in high-altitude adaptation. Likewise, patterns of amino acid variation at HBB-T1 and HBB-T2 are not consistent with divergent selection between high- and low-altitude populations. At HBB-T1 and HBB-T2, the s- and d-type alleles were present at roughly equal frequencies in the Lima and La Paz samples and levels of nucleotide differentiation were lower than those observed at the other polymorphic loci (Table 1). This result stands in stark contrast to patterns of α-globin variation in North American populations of deer mice, Peromyscus maniculatus. Deer mice possess three functionally distinct α-globin paralogs and each of the triplicated genes segregate functionally distinct alleles that exhibit pronounced allele frequency differences between high- and low-altitude populations (Snyder et al. 1988; Storz 2007; Storz et al. 2007). The physiological effects of α-globin variation in P. maniculatus contribute to fitness-related variation in aerobic performance (Chappell and Snyder 1984; Chappell et al. 1988; Hayes and O'connor 1999), and the abrupt altitudinal shifts in allele frequencies appear to be attributable to spatially varying selection that drives the divergent fine-tuning of hemoglobin function between different elevational zones (Snyder 1981; Snyder et al. 1988; Storz et al. 2007). Although North American deer mice and South American house mice inhabit a similarly broad range of altitudes, deer mice are indigenous to montane regions and have therefore had a much longer period of time to adapt to the physiological rigors of life at high altitude. By contrast, house mice are not commonly found in high-altitude environments in their native range, and their recent colonization of high-altitude environments in the Andes may not have provided enough time for selection to produce adaptive modifications of hemoglobin function. Alternatively, selection may have simply found a different solution to the physiological challenges associated with high-altitude hypoxia that did not involve biochemical modifications of blood oxygen affinity. The α-globin paralogs of deer mice and the β-globin paralogs of house mice both appear to represent cases of long-term balanced polymorphism, but in each case the allelic variation appears to be maintained by different modes of balancing selection.
Implications for the evolution of duplicated genes:
The fact that the two-locus Hbbs and Hbbd haplotypes are present at intermediate frequencies in house mouse populations throughout the world suggests a paradox regarding the fixation and subsequent retention of duplicated genes by natural selection. Theory suggests that the functional divergence of duplicated genes may typically be preceded and promoted by the adaptive maintenance of divergent alleles at the ancestral, single-copy gene (Spofford 1969; Walsh 2003; Proulx and Phillips 2006). The segregation load associated with overdominant selection can produce a substantial fitness advantage for newly arisen gene duplications because functional specialization of alternative alleles at the ancestral, single-copy gene is converted into a specialization of function between the two nascent paralogs (Spofford 1969; Proulx and Phillips 2006). If the duplication is preserved by selection that favors some form of physiological division of labor between the two functionally distinct paralogs, then the diversifying effects of this selection would be counteracted by the homogenizing effects of interparalog gene conversion. Thus, the fitness effects that promoted the original fixation and functional divergence of HBB-T1 and HBB-T2 should also favor the Hbbd haplotype (which possesses a pair of functionally distinct paralogs) over the Hbbs haplotype (which possesses a pair of identical paralogs). If the original duplication was fixed by positive selection, and if the Hbbd haplotype represents the ancestral condition, why is the Hbbs haplotype present at intermediate frequency in geographically disparate populations across the species' range? A resolution of this apparent paradox will require a thorough characterization of β-globin polymorphism in the ancestral range of M. musculus, in addition to functional studies to elucidate the causes of fitness variation among the alternative two-locus β-globin genotypes. Surveys of β-globin variation in Old World M. musculus and other closely related species of Mus may also help to unravel the complex history of recombination and interparalog gene conversion between HBB-T1 and HBB-T2.
Another question of relevance to the evolution of duplicated genes concerns the mechanism that has enabled the two β-globin paralogs on the Hbbd haplotype to escape from concerted evolution. Erhart et al. (1985) identified an extended stretch of simple-sequence repeats in intron 2 of HBB-T2 and suggested that this repeat array may promote the type of recombination event that results in gene conversion. This stretch of simple-sequence repeats is present on the Hbbs haplotype but has been deleted from the Hbbd haplotype. In fact, the location of this complex array of simple-sequence repeats corresponds to the region of unalignable sequence between nucleotide positions 827 and 1277 of HBB-T2. Erhart et al. (1985) argued that the deletion and/or disruption of this array of simple-sequence repeats helped produce a block of nonhomology between HBB-T1 and HBB-T2 on the Hbbd haplotype that inhibits interparalog gene conversion. According to the hypothesis of Erhart et al. (1985), this intronic block of nonhomology between HBB-T1 and HBB-T2 on the Hbbd haplotype provided a means of escape from concerted evolution and therefore facilitated functional divergence between the two paralogs.
Functional significance of amino acid polymorphism:
The fact that alternative β-globin protein alleles are consistently maintained at intermediate frequencies across the species' range suggests some form of balancing selection, and the strong LD between the two paralogs suggests the possibility that epistatic selection is maintaining coadapted combinations of alleles at HBB-T1 and HBB-T2. It is therefore important to determine the possible functional significance of the observed amino acid differences between the products of the s- and d-type β-globin alleles.
Studies of inbred strains and wild-caught mice have revealed that the β-chain products of the Hbbd haplotype are associated with a significantly higher hemoglobin-oxygen affinity than the products of the Hbbs haplotype (Newton and Peters 1983). Mice that were Hbbd/Hbbd homozygotes were also characterized by higher hemoglobin and hematocrit levels than Hbbs/Hbbd heterozygotes or Hbbs/Hbbs homozygotes. Since Hbbd/Hbbd mice are characterized by especially high blood oxygen affinities, Newton and Peters (1983) suggested that the higher hemoglobin and hematocrit levels in Hbbd/Hbbd homozygotes compensates for the reduced efficacy of tissue oxygenation. In an intensively studied island population of house mice in the North Atlantic, Berry and Murphy (1970) suggested that consistent seasonal shifts in β-globin genotype frequencies could be explained by selection in favor of Hbbd/Hbbd homozygotes during the overwintering period followed by overdominance of fitness or selection in favor of Hbbs/Hbbs homozygotes during the summer breeding months. Berry (1978) suggested that the fitness advantage of Hbbd/Hbbd mice during the overwintering period may be related to selection for increased thermogenic capacity during periods of severe cold stress. The fitness advantage of heterozygotes and/or Hbbs/Hbbs homozygotes during the summer breeding months is not clear.
It is also possible that the variation in hemoglobin-oxygen affinity is simply an indirect effect of genetically based variation in red blood cell metabolism. Mice carrying Hbbs haplotypes produce a single β-globin with one reactive cysteine residue, 93(F9)Cys, whereas mice carrying Hbbd haplotypes produce structurally distinct β-globins that each contain an additional reactive cysteine, 13(A10)Cys (Figure 3). These two additional reactive cysteine residues play a pivotal role in intraerythrocyte glutathione and NO metabolism (Miranda 2000; Giustarini et al. 2006; Hempe et al. 2007). As a result of allelic differences in the number of reactive β-chain sulfhydryl groups, inbred strains that are Hbbd/Hbbd homozygotes have higher intracellular concentrations of S-glutathionyl hemoglobin and S-nitrosohemoglobin than Hbbs/Hbbs homozygotes (Giustarini et al. 2006; Hempe et al. 2007). The intraerythrocyte fraction of S-glutathionyl hemoglobin determines the availability of reduced glutathione as a substrate for enzymatic detoxification reactions (Wu et al. 2004; Rahman et al. 2005) and S-nitrosohemoglobin transports NO used in vasoregulation (Pawloski et al. 2001; Mcmahon et al. 2005; Singel and Stamler 2005). In mice, genetically based variation in the concentration of reactive β-chain sulfhydryl groups influences intracellular reduction-oxidation balance and may therefore play a role in resistance to blood-borne pathogens (Giustarini et al. 2006; Hempe et al. 2007). Malaria parasites are highly sensitive to intraerythrocyte redox balance (Becker et al. 2004), and NO and oxygen radicals produced by macrophages play an essential role in host immune defense (Moncada et al. 1991). Interestingly, inbred strains of mice that are Hbbd/Hbbd homozygotes are generally more resistant to trypanosome infection and malaria infection by rodent plasmodia (Fortin et al. 2002; Duleu et al. 2004). Although a causal connection has yet to be established, the role of hemoglobin sulfhydryl groups on oxido-reductive reactions and blood-mediated metabolism of thiol reactants suggests the hypothesis that selection for resistance to blood-borne pathogens may play a role in the maintenance of β-globin polymorphism (Hempe et al. 2007), at least in some parts of the species' range.
Despite the well-documented functional differences between s- and d-type β-globin alleles, we cannot rule out the possibility that the unusually high nucleotide polymorphism and haplotype structure at house mouse β-globin genes is actually attributable to hitchhiking with a closely linked gene under balancing selection. In principle, a more extensive survey of LD across the β-globin gene cluster should reveal whether selection at linked sites is a plausible explanation for the observed patterns of variation.
Conclusion:
In South American house mice, remarkably high levels of nucleotide polymorphism and LD at two closely linked β-globin paralogs are attributable to the segregation of two highly distinct sequence haplotypes. Levels and patterns of nucleotide polymorphism suggest a complex history of diversity-enhancing selection that may be responsible for long-term maintenance of the alternative protein alleles. Hemoglobin tetramers that incorporate products of the alternative two-locus β-globin haplotypes are associated with pronounced differences in glutathione and NO metabolism in red blood cells, suggesting a possible mechanism of selection on hemoglobin function. Additional work is required to determine whether the β-globin genes are in fact the true targets of selection.
Acknowledgments
We thank M. Dean, A. Di Rienzo, N. Ferrand, and two anonymous reviewers for helpful comments on the manuscript. This work was funded by grants to J.F.S. from the National Science Foundation (DEB-0614342) and the Nebraska Research Council.
References
- Acin-Perez, R., M. P. Bayona-Bafaluy, P. Fernandez-Silva, R. Moreno-Loshuertos, A. Perez-Martos et al., 2004. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguileta, G., J. P. Bielawski and Z. H. Yang, 2006. Proposed standard nomenclature for the alpha- and beta-globin gene families. Genes Genet. Syst. 81: 367–371. [DOI] [PubMed] [Google Scholar]
- Auffray, J. C., F. Vanlerberghe and J. Brittondavidian, 1990. The house mouse progression in Eurasia: a paleontological and archaeozoological approach. Biol. J. Linn. Soc. 41: 13–25. [Google Scholar]
- Baines, J. F., and B. Harr, 2007. Reduced X-linked diversity in derived populations of house mice. Genetics 175: 1911–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Bafaluy, M. P., R. Acin-Perez, J. C. Mullikin, J. S. Park, R. Moreno-Loshuertos et al., 2003. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 31: 5349–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, K., L. Tilley, J. L. Vennerstrom, D. Roberts, S. Rogerson et al., 2004. Oxidative stress in malaria parasite-infected erythrocytes: host-parasite interactions. Int. J. Parasitol. 34: 163–189. [DOI] [PubMed] [Google Scholar]
- Bellamy, D., R. J. Berry, M. E. Jakobson, W. Z. Lidicker, J. Morgan et al., 1973. Ageing in an island population of the house mouse. Age Ageing 2: 235–250. [DOI] [PubMed] [Google Scholar]
- Bencowitz, H., P. Wagner and J. West, 1982. Effect of change in P50 on exercise tolerance at high altitude: a theoretical study. J. Appl. Physiol. 53: 1487–1495. [DOI] [PubMed] [Google Scholar]
- Berry, R. J., 1978. Genetic variation in wild house mice: where natural selection and history meet. Am. Sci. 66: 52–60. [PubMed] [Google Scholar]
- Berry, R. J., and M. E. Jakobson, 1975. Ecological genetics of an island population of the house mouse. J. Zool. 173: 341–354. [Google Scholar]
- Berry, R. J., and H. M. Murphy, 1970. The biochemical genetics of an island population of the house mouse. Proc. R. Soc. Lond. Ser. B 176: 87–103. [Google Scholar]
- Berry, R. J., and J. Peters, 1975. Macquarie Island house mice: a genetical isolate on a sub-Antarctic island. J. Zool. 176: 375–389. [Google Scholar]
- Berry, R. J., and J. Peters, 1977. Heterogeneous heterozygosities in Mus musculus populations. Proc. R. Soc. Lond. Ser. B 197: 485–503. [DOI] [PubMed] [Google Scholar]
- Berry, R. J., and J. Peters, 1981. Allozymic variation in house mouse populations, pp. 242–253 in Mammalian Population Genetics, edited by M. H. Smith and J. Joule. University of Georgia Press, Athens, GA.
- Berry, R. J., J. Peters and R. J. Van Aarde, 1978. Sub-antarctic house mice: colonization, survival and selection. J. Zool. 184: 127–141. [Google Scholar]
- Betran, E., J. Rozas, A. Navarro and A. Barbadilla, 1997. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics 146: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouverot, P., 1985. Adaptation to Altitude-Hypoxia in Vertebrates. Springer-Verlag, Berlin.
- Bullard, R. W., 1972. Vertebrates at altitude, pp. 209–225 in Physiological Adaptations: Desert and Mountain, edited by M. K. Yousef, S. M. Horvath and R. W. Bullard. Academic Press, New York.
- Chappell, M. A., and L. R. G. Snyder, 1984. Biochemical and physiological correlates of deer mouse α chain hemoglobin polymorphisms. Proc. Natl. Acad. Sci. USA 81: 5484–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell, M. A., J. P. Hayes and L. R. G. Snyder, 1988. Hemoglobin polymorphisms in deer mice (Peromyscus maniculatus): physiology of beta-globin variants and alpha-globin recombinants. Evolution 42: 681–688. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., 2006. Balancing selection and its effects on sequences in nearby genome regions. PloS Genet. 2: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depaulis, F., and M. Veuille, 1998. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 15: 1788–1790. [DOI] [PubMed] [Google Scholar]
- Dickerson, R. E., and I. Geis, 1983. Hemoglobin: Structure, Function, Evolution, and Pathology. Benjamin/Cummings Publishing, Menlo Park, CA.
- D'Surney, S. J., and R. A. Popp, 1992. Oxygen association-dissociation and stability analysis on mouse hemoglobins with mutant α- and β-globins. Genetics 132: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleu, S., P. Vincendeau, P. Courtois, S. Semballa, I. Lagroye et al., 2004. Mouse strain susceptibility to trypanosome infection: an arginase-dependent effect. J. Immunol. 172: 6298–6303. [DOI] [PubMed] [Google Scholar]
- Erhart, M. A., K. S. Simons and S. Weaver, 1985. Evolution of mouse β-globin genes: a recent gene conversion in the Hbbs haplotype. Mol. Biol. Evol. 2: 304–320. [DOI] [PubMed] [Google Scholar]
- Erhart, M. A., K. Piller and S. Weaver, 1987. Polymorphism and gene conversion in mouse α-globin haplotypes. Genetics 115: 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 1985. Confidence-limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Fortin, A., M. M. Stevenson and P. Gros, 2002. Susceptibility to malaria as a complex trait: big pressure from a tiny creature. Hum. Mol. Genet. 11: 2469–2478. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 1994. Alternatives to the neutral theory, pp. 1–17 in Non-Neutral Evolution: Theories and Molecular Data, edited by B. Golding. Chapman & Hall, New York.
- Gilman, J. G., 1976. Mouse hemoglobin beta chains: comparative sequence data on adult major and minor beta chain from two species. Biochem. J. 159: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustarini, D., I. Dalle-Donne, E. Cavarra, S. Fineschi, G. Lungarella et al., 2006. Metabolism of oxidants by blood from different mouse strains. Biochem. Pharmacol. 71: 1753–1764. [DOI] [PubMed] [Google Scholar]
- Goodman, M., 1986. Molecular evidence on the ape subfamily hominidae, pp. 121–132 in Evolutionary Perspectives in the New Genetics, edited by H. Gershowitz, R. D. Rugknagel and R. E. Tashian. A. R. Liss, New York.
- Guenet, J. L., and F. Bonhomme, 2003. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 19: 24–31. [DOI] [PubMed] [Google Scholar]
- Harr, B., 2006. Genomic islands of differentiation between house mouse subspecies. Genome Res. 16: 730–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, J. P., 1989. Altitudinal and seasonal effects on aerobic metabolism of deer mice. J. Comp. Physiol. B, Biochem. Syst. Environ. Physiol. 159: 453–459. [DOI] [PubMed] [Google Scholar]
- Hayes, J. P., and C. S. O'Connor, 1999. Natural selection on thermogenic capacity of high-altitude deer mice. Evolution 53: 1280–1287. [DOI] [PubMed] [Google Scholar]
- Hempe, J. M., J. Ory-Ascani and D. Hsia, 2007. Genetic variation in mouse beta globin cysteine content modifies glutathione metabolism: implications for the use of mouse models. Exp. Biol. Med. 232: 437–444. [PubMed] [Google Scholar]
- Hey, J., and J. Wakeley, 1997. A coalescent estimator of the population recombination rate. Genetics 145: 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilse, K., and R. A. Popp, 1968. Gene duplication as the basis for amino acid ambiguity in the alpha-chain polypeptides of mouse hemoglobins. Proc. Natl. Acad. Sci. USA 61: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochachka, P. W., and G. N. Somero, 2002. Biochemical Adaptation. Oxford University Press, New York.
- Hudson, R. R., 2000. A new statistic for detecting genetic differentiation. Genetics 155: 2011–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1988. The coalescent process in models with selection and recombination. Genetics 120: 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguade, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., D. D. Boos and N. L. Kaplan, 1992. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 9: 138–151. [DOI] [PubMed] [Google Scholar]
- Hutton, J. J., J. Bishop, R. Schweet and E. S. Russell, 1962. Hemoglobin inheritance in inbred mouse strains. I. Structural differences. Proc. Natl. Acad. Sci. USA 48: 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideraabdullah, F. Y., E. de la Casa-Esperon, T. A. Bell, D. A. Detwiler, T. Magnuson et al., 2004. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome Res. 14: 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., 2003. The coalescent and infinite-site model of a small multigene family. Genetics 163: 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, L., and D. Pilbeam, 1980. Of mice and men: fossil-based divergence dates and molecular “clocks”. J. Hum. Evol. 9: 551–555. [Google Scholar]
- Jobb, G., A. von Haeseler and K. Strimmer, 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kelly, J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, J. K., and M. J. Wade, 2000. Molecular evolution near a two-locus balanced polymorphism. J. Theor. Biol. 204: 83–101. [DOI] [PubMed] [Google Scholar]
- Kimura, M., and T. Ohta, 1973. Age of a neutral mutant persisting in a finite population. Genetics 75: 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Leder, P., J. N. Hansen, D. Konkel, A. Leder, Y. Nishioka et al., 1980. Mouse globin system: a functional and evolutionary analysis. Science 209: 1336–1342. [DOI] [PubMed] [Google Scholar]
- Lenfant, C., 1973. High altitude adaptation in mammals. Am. Zool. 13: 447–456. [Google Scholar]
- Lundrigan, B. L., S. A. Jansa and P. K. Tucker, 2002. Phylogenetic relationships in the genus Mus, based on paternally, maternally, and biparentally inherited characters. Syst. Biol. 51: 410–431. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- McMahon, T. J., G. S. Ahearn, M. P. Moya, A. J. Gow, Y. C. T. Huang et al., 2005. A nitric oxide processing defect of red blood cells created by hypoxia: deficiency of S-nitrosohemoglobin in pulmonary hypertension. Proc. Natl. Acad. Sci. USA 102: 14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, J. J., 2000. Highly reactive cysteine residues in rodent hemoglobins. Biochem. Biophys. Res. Commun. 275: 517–523. [DOI] [PubMed] [Google Scholar]
- Moncada, S., R. M. J. Palmer and E. A. Higgs, 1991. Nitric-oxide: physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 43: 109–142. [PubMed] [Google Scholar]
- Monge, C., and F. León-Velarde, 1991. Physiological adaptation to high altitude: oxygen transport in mammals and birds. Physiol. Rev. 71: 1135–1172. [DOI] [PubMed] [Google Scholar]
- Myers, J. H., 1974. Genetic and social structure of feral house mouse populations on Grizzly Island, California. Ecology 55: 747–759. [Google Scholar]
- Nachman, M. W., 1997. Patterns of DNA variability at X-linked loci in Mus domesticus. Genetics 147: 1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman, M. W., S. N. Boyer, J. B. Searle and C. F. Aquadro, 1994. Mitochondrial DNA variation and the evolution of Robertsonian chromosomal races of house mice. Genetics 136: 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, A., and N. H. Barton, 2002. The effects of multilocus balancing selection on neutral variability. Genetics 161: 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and W. -H. Li, 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, M. F., and J. Peters, 1983. Physiological variation of mouse haemoglobin. Proc. R. Soc. Lond. Ser. B 218: 443–453. [DOI] [PubMed] [Google Scholar]
- Nishioka, Y., and P. Leder, 1979. The complete sequence of a chromosomal mouse α-globin gene reveals elements conserved throughout vertebrate evolution. Cell 18: 875–882. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., and H. Innan, 2003. The genealogy of sequences containing multiple sites subject to strong selection in a subdivided population. Genetics 163: 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawloski, J. R., D. T. Hess and J. S. Stamler, 2001. Export by red blood cells of nitric oxide bioactivity. Nature 409: 622–626. [DOI] [PubMed] [Google Scholar]
- Perutz, M. F., 1983. Species adaptation in a protein molecule. Mol. Biol. Evol. 1: 1–28. [DOI] [PubMed] [Google Scholar]
- Petras, M., 1967. Studies of natural populations of Mus. I. Biochemical polymorphisms and their bearing on breeding structure. Evolution 21: 259–274. [DOI] [PubMed] [Google Scholar]
- Petras, M. L., and J. C. Topping, 1983. The maintenance of polymorphisms at two loci in house mouse (Mus musculus) populations. Can. J. Genet. Cytol. 25: 190–201. [DOI] [PubMed] [Google Scholar]
- Pfunder, M., O. Holzgang and J. E. Frey, 2004. Development of microarray-based diagnostics of voles and shrews for use in biodiversity monitoring studies, and evaluation of mitochondrial cytochrome oxidase I vs. cytochrome b as genetic markers. Mol. Ecol. 13: 1277–1286. [DOI] [PubMed] [Google Scholar]
- Popp, R. A., 1973. Sequence of amino acids in the β-chain of single hemoglobins from C57BL, SWR, and NB mice. Biochim. Biophys. Acta 303: 52–60. [DOI] [PubMed] [Google Scholar]
- Popp, R. A., E. G. Bailiff, L. C. Skow and J. B. I. Whitney, 1982. The primary structure of genetic variants of mouse hemoglobin. Biochem. Genet. 20: 199–208. [DOI] [PubMed] [Google Scholar]
- Poyart, C., H. Wajcman and J. Kister, 1992. Molecular adaptation of hemoglobin-function in mammals. Respir. Physiol. 90: 3–17. [DOI] [PubMed] [Google Scholar]
- Prager, E. M., R. D. Sage, U. Gyllensten, W. K. Thomas, R. Hubner et al., 1993. Mitochondrial-DNA sequence diversity and the colonization of Scandinavia by house mice from East Holsten. Biol. J. Linn. Soc. 50: 85–122. [Google Scholar]
- Prager, E. M., C. Orrego and R. D. Sage, 1998. Genetic variation and phylogeography of central Asian and other house mice, including a major new mitochondrial lineage in Yemen. Genetics 150: 835–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx, S. R., and P. C. Phillips, 2006. Allelic divergence precedes and promotes gene duplication. Evolution 60: 881–892. [PubMed] [Google Scholar]
- Rahman, I., S. K. Biswas, L. A. Jimenez, M. Torres and H. J. Forman, 2005. Glutathione, stress responses, and redox signaling in lung inflammation. Antioxid. Redox Signal. 7: 42–59. [DOI] [PubMed] [Google Scholar]
- Raymond, M., and F. Rousset, 1995. An exact test for population differentiation. Evolution 49: 1280–1283. [DOI] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Sabeti, P. C., D. E. Reich, J. M. Higgins, H. Z. P. Levine, D. J. Richter et al., 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- Sage, R. D., 1981. Wild mice, pp. 39–90 in The Mouse in Biomedical Research: History, Genetics, and Wild Mice, edited by H. L. Foster, J. D. Small and J. G. Fox. Academic Press, New York.
- Sage, R. D., J. B. I. Whitney and A. C. Wilson, 1986. Genetic Analysis of a Hybrid Zone Between Domesticus and Musculus Mice (Mus Musculus Complex): Hemoglobin Polymorphisms. Springer-Verlag, New York. [DOI] [PubMed]
- Selander, R. K., and S. Y. Yang, 1969. Protein polymorphism and genic heterozygosity in a wild population of the house mouse (Mus musculus). Genetics 63: 653–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander, R. K., W. G. Hunt and S. Y. Yang, 1969. a Protein polymorphism and genic heterozygosity in two European subspecies of the house mouse. Evolution 23: 379–390. [DOI] [PubMed] [Google Scholar]
- Selander, R. K., S. Y. Yang and W. G. Hunt, 1969. b Polymorphism in esterases and hemoglobin in wild populations of the house mouse (Mus musculus). Univ. Texas Publ. 6918: 271–338. [Google Scholar]
- She, J. X., F. Bonhomme, P. Boursot, L. Thaler and F. Catzeflis, 1990. Molecular phylogenies in the genus Mus: comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol. J. Linn. Soc. 41: 83–103. [Google Scholar]
- Singel, D. J., and J. S. Stamler, 2005. Chemical physiology of blood flow regulation by red blood cells: the role of nitric oxide and S-nitrosohemoglobin. Ann. Rev. Physiol. 67: 99–145. [DOI] [PubMed] [Google Scholar]
- Slatkin, M., 2000. Balancing selection at closely linked, overdominant loci in a finite population. Genetics 154: 1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, L. R. G., 1981. Deer mouse hemoglobins: Is there genetic adaptation to high altitude? Bioscience 31: 299–304. [Google Scholar]
- Snyder, L. R. G., M. A. Chappell and J. P. Hayes, 1988. α-Chain hemoglobin polymorphisms are correlated with altitude in the deer mouse, Peromyscus maniculatus. Evolution 42: 689–697. [DOI] [PubMed] [Google Scholar]
- Spofford, J. B., 1969. Heterosis and the evolution of duplications. Am. Nat. 103: 407–432. [Google Scholar]
- Springer, M. K., W. J. Murphy, E. Eizirik and S. J. O'Brien, 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. USA 100: 1056–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, J. F., 2007. Hemoglobin function and physiological adaptation to hypoxia in high-altitude mammals. J. Mammal. 88: 24–31. [Google Scholar]
- Storz, J. F., S. J. Sabatino, F. G. Hoffmann, E. J. Gering, H. Moriyama et al., 2007. The molecular basis of high-altitude adaptation in deer mice. PloS Genet. 3: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobeck, C., 1983. Expected linkage disequilibrium for a neutral locus linked to a chromosomal rearrangement. Genetics 103: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., T. Shimada, M. Terashima, K. Tsuchiya and K. Aplin, 2004. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 33: 626–646. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, A., Y. H. Liu and N. Saitou, 2004. Genetic variation versus recombination rate in a structured population of mice. Mol. Biol. Evol. 21: 404–409. [DOI] [PubMed] [Google Scholar]
- Terashima, M., S. Furusawa, N. Hanzawa, K. Tsuchiya, A. Suyanto et al., 2006. Phylogeographic origin of Hokkaido house mice (Mus musculus) as indicated by genetic markers with maternal, paternal and biparental inheritance. Heredity 96: 128–138. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian, C., R. S. Ajioka, L. B. Jorde, J. P. Kushner and M. Kreitman, 2003. A method for detecting recent selection in the human genome from allele age estimates. Genetics 165: 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, P. K., B. K. Lee and E. M. Eicher, 1989. Y-chromosome evolution in the subgenus Mus (genus Mus). Genetics 122: 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek, Z., F. Kreuzer and L. Hoofd, 1973. Advantage or disadvantage of a decrease of blood oxygen affinity for tissue oxygen supply at hypoxia. A theoretical study comparing man and rat. Pflügers Arch. 342: 185–187. [DOI] [PubMed] [Google Scholar]
- Walsh, B., 2003. Population-genetic models of the fates of duplicate genes. Genetica 118: 279–294. [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Wheeler, L. L., and R. K. Selander, 1972. Genetic variation in populations of the house mouse, Mus musculus, in the Hawaiian Islands. Univ. Texas Publ. 7213: 269–296. [Google Scholar]
- Whitney, J. B. I., 1978. Simplified typing of mouse hemoglobin (Hbb) phenotypes using cystamine. Biochem. Genet. 16: 667–672. [DOI] [PubMed] [Google Scholar]
- Whitney, J. B. I., G. T. Copland, L. C. Skow and E. S. Russell, 1979. Resolution of products of the duplicated hemoglobin α-chain loci by isoelectric focusing. Proc. Natl. Acad. Sci. USA 76: 867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, J. B. I., R. R. Cobb, R. A. Popp and T. W. O'Rourke, 1985. Detection of neutral amino acid substitutions in proteins. Proc. Natl. Acad. Sci. USA 82: 7646–7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf, C., K. Zhao, H. Innan and M. Nordborg, 2004. The probability and chromosomal extent of trans-specific polymorphism. Genetics 168: 2363–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., Y. Z. Fang, S. Yang, J. R. Lupton and N. D. Turner, 2004. Glutathione metabolism and its implications for health. J. Nutr. 134: 489–492. [DOI] [PubMed] [Google Scholar]