Abstract
Understanding the origin and evolution of sex chromosomes requires studying recently evolved X–Y chromosome systems such as those in some flowering plants. We describe Y chromosome deletion mutants of Silene latifolia, a dioecious plant with heteromorphic sex chromosomes. The combination of results from new and previously described deletions with histological descriptions of their stamen development defects indicates the presence of two distinct Y regions containing loci with indispensable roles in male reproduction. We determined their positions relative to the two main sex determination functions (female suppressing and the other male promoting). A region proximal to the centromere on the Y p arm containing the putative stamen promoting sex determination locus includes additional early stamen developmental factors. A medial region of the Y q arm carries late pollen fertility factors. Cytological analysis of meiotic X–Y pairing in one of the male-sterile mutants indicates that the Y carries sequences or functions specifically affecting sex chromosome pairing.
IT is widely accepted that X–Y sex chromosome systems evolved from a pair of autosomes, which initially acquired the mutation(s) responsible for sexual dimorphism, the primary sex determination genes (reviewed in Charlesworth 1991). Two main processes are generally considered to occur during this earliest stage of X–Y evolution. One is restriction of crossing over in the region(s) containing these loci, which might be called a proto-X–Y system. The second is the accumulation of two kinds of genes advantageous for male sex functions: sexually antagonistic male-benefit alleles and alleles enhancing male gamete success.
Sexually antagonistic male-benefit alleles are predicted to accumulate close to the sex-determining genes, because expression of their disadvantageous effects in females will be reduced by tight linkage with the sex-determining region (Fisher 1931; Rice 1987; reviewed in Vicoso and Charlesworth 2006). Recombination between the primary sex-determining region and loci at which male-benefit alleles have accumulated on the Y will generate poorly adapted phenotypes, selecting for tighter linkage in the region. The process of selection and recruitment of male-benefit alleles may be followed by further selection favoring restriction of recombination, which may eventually be completely suppressed, and the entire proto-Y-chromosome region may become nonrecombining. This can explain why, in many sex chromosome systems, recombination is suppressed across most of the Y, not just in the primary sex-determining region (reviewed in Charlesworth et al. 2005). In mammals, for example, only small parts of the Y (the “pseudo-autosomal” regions, PAR1 and PAR2; see Perry and Ashworth 1999; Charchar et al. 2003) still recombine with the X.
After crossing over ceases, the sequences of the initially homologous regions of the proto-sex chromosomes diverge, both due to further mutations in genes linked to the sex-determining region, as just outlined, and to substitutions of neutral and weakly deleterious mutations under genetic drift. The low effective population size of the evolving Y chromosome also leads to genetic degeneration within the nonrecombining sex-specific region, including reduction of genes' functions, loss of genes, and accumulation of transposable elements (reviewed in Charlesworth and Charlesworth 2000).
Empirical evidence for the involvement of sexually antagonistic male-benefit alleles and for their presence on the Y driving the evolution of low crossing over is limited. The classical evidence for accumulation of male-benefit alleles closely linked to sex-determining genes is the presence, in the fish Poecilia reticulata, of male sexual attractiveness alleles in both the nonrecombining and pseudo-autosomal regions of the Y chromosome (Winge 1927; Bull 1983; Lindholm and Breden 2002). These alleles reduce males' survival to maturity, even under laboratory conditions (Brooks 2000), so they are clearly disadvantageous, except for their effects on male sexual attractiveness. They are probably not sex limited in expression (Bull 1983), but the benefits of attractiveness in females are less important than for males, and there are fitness costs of ornamentation in terms of conspicuousness to predators; thus, these are probably sexually antagonistic genes of the kind hypothesized. There is also some quantitative genetic support for sexually antagonistic alleles in Drosophila melanogaster (Gibson et al. 2002), and there is indirect evidence in the neo-Y system of D. americana (Mcallister and Evans 2006). Recently, a QTL mapping study inferred the presence of sexually antagonistic genes for sexually dimorphic characters in Silene latifolia, including flower size (Scotti and Delph 2006). However, these conclusions need to be verified by other kinds of markers, as the genes were inferred to be located in two PAR regions (which has not previously been suspected). Moreover, the situation is not the one hypothesized above: females apparently do not express the phenotypes (expression is sex limited), and the genes were hypothesized to be maintained polymorphic by antagonistic pleiotropy in males.
An indirect line of evidence is based on the human Y, the best-studied Y chromosome so far. As the theory above predicts, the Y sequence has revealed numerous genes involved in spermatogenesis. Some of these are genes that were once autosomal or X-linked and have transposed onto the Y (Lahn and Page 1999b; Skaletsky et al. 2003), while others are multi-copy genes, present in ampliconic regions, mostly with no homology with either X-linked or autosomal genes (Skaletsky et al. 2003). The human Y is thus enriched for male function genes, many of which are indispensable for male reproduction (their loss leads to oligospermia or azoospermia, disorders that disallow reproduction under natural conditions (these data are reviewed in Kent-First 2000; Ellis and Affara 2006).
There is evidence that recombination arrest between the mammalian sex chromosomes evolved in successive stages, as the above model predicts. This is deduced from the observation of X chromosome evolutionary strata. The strata are genomic regions with different divergence between homologous X and Y gene pairs, with the least diverged region (indicating the most recent recombination arrest) located closest to the PAR1 on the genetic map of the X, and the most diverged (in which recombination between the X and Y ceased longest ago) furthest from the PAR1 (Lahn and Page 1999a; Sandstedt and Tucker 2004). The gene order of mammalian X chromosomes is highly conserved, whereas the Y chromosomes are very rearranged, which suggests that inversions have occurred on the Y during these chromosomes' history, estimated to be a period of ∼300 million years (Lahn and Page 1999a). The divergence values are not discontinuous enough to define boundaries definitively indicating inversions, so loss of crossing over may have involved rearrangements and/or gradual recombination arrest (Iwase et al. 2003; Marais and Galtier 2003; Skaletsky et al. 2003; Ross et al. 2005).
Mammalian X–Y systems are ancient and thus not suited to gaining a detailed understanding of the early stages of this evolution or for testing the sexual antagonism hypothesis. The recent evolutionary history of flowering plants (Leebens-Mack et al. 2005), and the scattered distribution of dioecy (individuals with separate sexes) among them, suggest recent evolution of dioecy in angiosperms, sometimes even within plant genera, such as the genus Silene (Desfeux et al. 1996), yet some dioecious plants have X–Y sex chromosome systems (Westergaard 1958; Ainsworth et al. 1998; Charlesworth 2002). Early events in X–Y evolution can thus be studied in plants (reviewed in Negrutiu et al. 2001; Vyskot and Hobza 2004). Dioecious species of the genus Silene have started to be studied in detail. Their sex chromosomes are estimated to have evolved recently, only ∼10–20 MYA (Nicolas et al. 2005; Bergero et al. 2007). The available evidence suggests that these Y chromosomes are not highly degenerated; the DNA is not hypermethylated (Siroky et al. 1998), nor is there histone H4 hypoacetylation compared with the X (Vyskot et al. 1999). The nonrecombining Y region includes most of the length of the Y, which is longer than the X, and a PAR region is present on the q arm (Lengerova et al. 2003). Partial deletions of the S. latifolia Y chromosome have revealed three regions controlling different sex functions. One region suppresses female sex organ development (deletion strains have hermaphrodite flowers; Westergaard 1946; Lardon et al. 1999), while deletions of two other regions lead to anther arrest (deletion strains bear asexual flowers in which male structures closely resemble those in females; Donnison et al. 1996; Farbos et al. 1999) or late male sterility (Westergaard 1946), showing that these regions carry genes involved in male sex organ development and male fertility, respectively. The generally accepted cytogenetic model (Figure 1) is that the regions corresponding to hermaphroditic and asexual phenotypes contain the two sex determination functions that are responsible for the sexual dimorphism in this species.
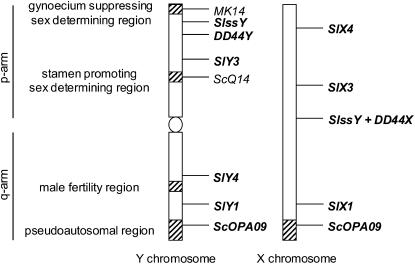
Figure 1.—
The S. latifolia sex chromosomes and the current sex determination model according to Negrutiu et al. (2001) and Charlesworth (2002). Y-chromosome deletion mutants (Westergaard 1958; Donnison et al. 1996; Farbos et al. 1999; Lardon et al. 1999; Lebel-Hardenack et al. 2002; Zluvova et al. 2005a) define at least two critical sex-determining regions on the differential arm of the Y-chromosome: the regions responsible for suppressing female organ development and for early stamen development (see text), while the arm homologous with the X (i.e., containing the PAR) carries a region responsible for late stamen development (plants lacking this region are male sterile). When compared to the known gene order on the X chromosome, the presence of an inversion can be deduced. Molecular markers are listed on the right, those present both on the X and Y chromosomes are in bold. The p and q arms are indicated.
More detailed dissection and localization of different functional regions of the Y chromosome is now possible with the help of molecular markers. Sex-linked markers in S. latifolia (supplemental Table S1 at http://www.genetics.org/supplemental/; Figure 1) have been used to estimate the recombination map of the X and relate this to estimated X–Y sequence divergence (Filatov 2005a; Nicolas et al. 2005; Bergero et al. 2007) and to compare the X and Y gene orders using deletion maps of the Y (Lebel-Hardenack et al. 2002; Zluvova et al. 2005a). They have also been used to understand the origins of the sex chromosome pair and whether translocations from other chromosomes have been involved (Filatov 2005a; Markova et al. 2006). These results suggest that, similarly to the mammalian sex chromosomes, restriction of recombination between the S. latifolia X and Y evolved gradually (Filatov 2005a; Nicolas et al. 2005; Bergero et al. 2007). They further suggested that a large inversion inferred on the Y chromosome may have caused the observed rearranged order of genes but probably did not cause X–Y recombination arrest (Zluvova et al. 2005a), since the genes within the inverted region include pairs with a wide range of X–Y silent site divergence values.
Here, we study a set of 16 S. latifolia Y chromosome deletions described previously (Farbos et al. 1999; Lardon et al. 1999) plus 12 new deletions. For each deletion that affects male functions, we describe the anther and pollen phenotypic defects in detail to estimate the number of genes carried on the Y chromosome whose loss abolishes male functions. These could be genes in two categories: (i) genes with copies on the X whose X alleles cannot replace the Y gene functions (suggesting that the Y allele has become more male-promoting, or that the X carries an allele that has weakened in male function; among the latter will be the gene in which a mutation caused the appearance of females in the species); and (ii) genes with no X copies, such as the Y-linked genes in Drosophila (Carvalho 2002).
We also tested each mutant for the presence or absence of molecular markers to find out which of them are due to deletion of Y chromosomal regions. The mapping work revealed no inconsistencies with previous maps of the Y and largely confirmed previous conclusions. This allowed us to base our conclusions on the best ordering of the deletions so far possible, as well as to demonstrate Y linkage of the new mutants studied. Finally, we examined X–Y chromosome meiotic pairing in all the mutants and identified three Y deletion mutants exhibiting altered X–Y pairing during meiosis.
MATERIALS AND METHODS
Rationale and methodology of the screen:
The results in this article are based on screening deletion mutants obtained by irradiating pollen grains, pollinating wild-type females with irradiated pollen, and screening for abnormalities in the M1 generation. Recessive mutations on the autosomes or the X chromosome should be masked by their wild-type alleles in the maternal genome, whereas dominant or dosage-sensitive mutations in autosomal and X-linked genes will be detected in the screen. Plants with deletions of Y-linked genes that do not have functional homologs within the genome (type i in the list above) will also show phenotypic defects, as will plants with mutations in genes of type ii listed above, but recessive mutations in Y-linked genes with functional homologs elsewhere in the genome will show no phenotypic defects. Y chromosome deletions can be distinguished from autosomal and X-linked mutations by using Y-linked markers. Thus, although a large number of genes may exist on the S. latifolia Y chromosome, our screen should detect a specific category of interest: those with no functional homologs within the genome.
One mutant (pf5, see below) arose spontaneously in a family grown from a cross between plants from two laboratory strains provided by M. Ruddat (University of Chicago). All of the other mutants were generated by gamma-irradiation of pollen from a single male plant, which was used to pollinate a single female parent (Lardon et al. 1999). The mutant plants were vegetatively propagated, and flower buds of several replicate individuals were analyzed for each deletion.
Y-specific markers were used to confirm the presence of Y deletions and to localize them on the previously published S. latifolia Y deletion map (Zluvova et al. 2005a; Hobza et al. 2006). Details of the markers are shown in supplemental Table S1 (at http://www.genetics.org/supplemental/), which includes references to the publications in which the conditions for the PCR amplifications are described. All the markers that we genotyped in the mutants were completely Y-linked.
Cytological and histological analysis:
Mitotic and meiotic preparations were performed as described in Lardon et al. (1999). Flower bud fixation, paraplast embedding, histological preparations, and staining were performed as described previously (Zluvova et al. 2005b). Preparations were observed using an Olympus Provis AX70 microscope, and pictures were captured using ISIS (Metasystems, Sandhausen, Germany).
RESULTS
Generation of Y deletion mutants with sex phenotypes:
We expected our screen to recover mainly dominant or partially dominant mutations due to deletions or mutations of Y-linked genes that have no functional homologs elsewhere in the genome (see materials and methods), i.e., either null mutations or deletions (which are expected to behave as nulls). The screen of 2262 M1 plants identified a total of 36 plants with sex function mutations. Sixteen were described previously (14 hermaphroditic bsx mutants, 1 of them autosomal, and 2 asexual asx mutants; Farbos et al. 1999; Lardon et al. 1999). Here, we describe for the first time 12 stamen mutants (out of the remaining 20 mutants with stamen defects), for which we also demonstrate Y linkage below. No other morphological defects in floral traits, other than one floral homeotic and meristic mutation (Scutt et al. 1999) or the occasional albino, dwarf, or narrow-leaved plantlets, were observed (Lardon et al. 1999). Thus, male defects were found disproportionately compared with other phenotypes. This is consistent with these mutations being due to Y chromosome partial deletions.
Here, we characterize the male defects of the 12 new deletions and add more details to the descriptions of those previously studied. Our new histological analyses disclosed defects in a broad range of anther developmental processes. They include mutants with alterations in the early formation of the anthers, referred to as anther development mutations (ad1–7), two of which—denoted here by ad6 and ad7—were preliminarily described as “misparieta” mutants (msp1 and 2 in Hinnisdaels et al. 1997), and in microspore maturation (pollen fertility mutations, denoted by pf1–5). The bsx mutants are almost all male and female fertile (Lardon et al. 1999), but all mutants in the other categories above (asx, ad, and pf) are male sterile. The mutants have maintained their distinctive phenotypes when repeatedly propagated through cuttings (at least 10 vegetative clones per mutant in the last 10 years for all mutant subgroups in the asx, ad, and pf classes).
Meiotic behavior of mutants with stamen and pollen defects:
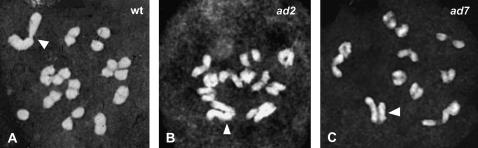
In most mutants, meiotic X–Y pairing was unchanged (Figure 2A), but abnormalities were seen in three independent lines. Two mutants, ad2 and ad3, showed variable rates of interstitial X–Y pairing, suggesting an inversion including the PAR region (Figure 2B), while ad7 exhibited a high frequency of X–Y pairing (84%), including extensive pairing in X–Y regions that never normally pair (Figure 2C).
Figure 2.—
Meiotic preparations of wild type and two Y-deletion mutants showing altered X–Y behavior in meiosis. Wild-type diakinesis. Note the terminal pairing of X and Y (A). Diakinesis in ad2 mutant in which X and Y do not pair. Note that the distal regions of the X are not aligned with any of the Y distal regions (B). Diakinesis in ad7 showing pairing of X and Y. One distal region of each chromosome is aligned (C). White arrowheads indicate X–Y chromosome pairs.
Deletions of Y chromosome markers:
The mutations are inferred to be due to partial deletions of the Y chromosome (see below), and, in some mutants, deletions are cytologically visible (Table 1). The largest deletion causing loss of male functions, asx1, lacks ∼40% of the p arm of the Y (Table 1) (Farbos et al. 1999). Most of the p arm (the arm that does not pair with the X) is covered by deletions detected in our screen. Deletions leading to hermaphrodite mutants have previously been estimated to extend to as much as 50% of the wild-type Y chromosome p arm (the largest is the bsx1 mutant reported by Lardon et al. 1999). Since these plants are largely male fertile, genes controlling stamen development and pollen fertility functions cannot be located in the part of the Y covered by these deletions.
TABLE 1.
Cytological characterization of male-defective mutants of S. latifolia
| Size of deletion (%) | Meiosisa
|
||||
|---|---|---|---|---|---|
| Mutant names | Diakinesis | Metaphase I | X–Y pairing | Other changes | |
| ad1 | 9 | 12 | 12 | Wild type | |
| ad2 | 12 | 12 | 12 | No or interstitial (36%) | |
| ad3 | Not measurable | 12 | 12 | Interstitial (11%) | 10% abnormal segregation in meiosis II |
| ad4 | 13 | 12 | 12 | Wild type | |
| ad5 | Not measurable | 12 | 12 | Wild type | |
| pf1 | 4 | 12 | 12 | Wild type | |
| pf2 | Not measurable | Not studied | Not studied | Wild type | |
| pf3 | Not measurable | 12 | 12 | Wild type | |
| pf4 | Not measurable | 12 and 13 (10%) | Mitotic divisions of one bivalent | Wild type | |
| pf5 | Not measurable | 14 | Wild type | ||
| ad6 (msp1) | 8 | 12 | 12 | Wild type | |
| ad7 (msp2) | 7 | 12 | 12 | Extensive X–Y pairing (84%) | |
| asx1 | 20 | No meiosis | Not testable | ||
| asx2 | 12 | No meiosis | Not testable | ||
The second column of the table shows the sizes of the Y deletions measured in mitotic cell preparations, as a percentage of the entire Y chromosome. An extensive meiotic analysis was performed to estimate bivalent formation during diakinesis and metaphase I (columns 3 and 4), other meiotic aberrations (column 5), and X–Y pairing behavior (column 6).
See Lardon et al. (1999): number of bivalents are known to vary from 2n = 12 to 2n = 14 in S. latifolia genotypes, with no consequences for the sex phenotype.
We tested each mutant strain for the presence of 13 previously described Y-linked markers, 6 genic and 7 anonymous (supplemental Table S1 at http://www.genetics.org/supplemental/). The 2 asx mutants, and the 12 pf and ad mutants (6 of which have no cytologically detectable deletions), were found to lack at least 1 of these markers, confirming that these involve deletions of parts of the Y chromosome. Figure 3 outlines the deletions found in the mutants.
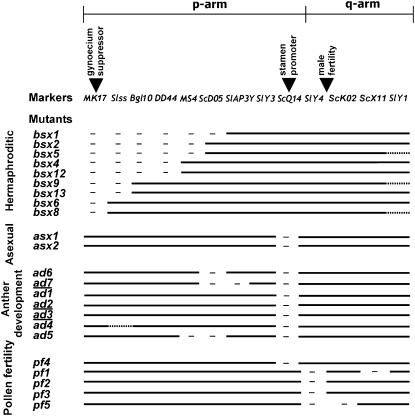
Figure 3.—
Outline of the deletions observed in the mutants analyzed. The markers are shown at the top, ordered according to the Y map (Hobza et al. 2006), with inverted triangles indicating the sex determination functions located on the p arm and a pollen fertility region on the q arm. The pseudo-autosomal region is on the right. The mutant names are given on the left. The underlined mutant names state for mutants with pairing phenotypes. Full lines indicate markers present in each deletion mutant, dashes indicate markers that were determined to be missing, and dotted lines indicate that the presence of the marker was not tested in the strain. Deletions are not to scale. Note that in several mutants more than one marker is deleted.
Histological characterization of the mutants:
The histological results for the asx, ad, and pf mutants are summarized in Table 2, and a more detailed description of mutants lacking one or more markers is given below and in supplemental Figures S1 and S2 (at http://www.genetics.org/supplemental/). The histological analyses of anthers define a variety of clear-cut male sex defects. The first two major categories, asexuals (asx) and anther developmental mutants (ad), have early anther developmental defects, while the third category includes pollen fertility mutants (pf). With detailed phenotypic data, these categories subdivide into a total of nine groups.
TABLE 2.
Outline description of the phenotypes of the deletion mutants and suggested function(s) of the genes involved
| Mutant class and Y deletion sizes | No. of deletion mutants | Mutant group names and possible gene functions involved in each subgroup (Roman numerals) | Phenotypic defects | Figures |
|---|---|---|---|---|
| Asexual; interstitial, up to 40% of proximal Yp | 2 | asx1, asx2 specifying primary parietal cell fate | Anther arrest before differentiation of the parietal layers, as in wild-type females. The two mutants have an identical phenotype. | 4B, S1B |
| Anther development; interstitial, up to 26% of proximal Yp | 7 | ad1–ad7 | Anthers arrested early during formation of parietal and sporogenous layers. Completely sterile. | |
| i. ad1 stamen sub-whorl coordination | Inner, antipetalous, anther subwhorl is arrested at the four-lobed stage, deposits accumulate around the connective region. Outer, antisepalous anthers form abnormally small and unviable pollen grains. | 4C, 4Q, S2K, S2L | ||
| ii. ad2, ad6 maintaining parietal layer | Anther arrest at the stage of parietal layer formation in the wild type, but microspore mother cells can undergo meiosis. The two mutants have very similar phenotype. | 4E, S1E, S1D | ||
| iii. ad3, ad5 locule synchronization | Asymmetrically developed anthers (abaxial locules form only primary parietal layers). The two mutants have an identical phenotype. | 4F, S1G, S1H | ||
| iv. ad4, ad7 specification of parietal layers | Secondary parietal layer often differentiated directly into tapetum-like cells without formation of all anther layers. The two mutants have very similar phenotype. Ad7 is altered in X–Y pairing. | 4H, S1J, S1K | ||
| Pollen fertility; interstitial, up to 8% of proximal or medial Yq | 5 | pf1–pf5 | Microspores either degenerate after meiosis, or the pollen grains are sterile. | |
| i. pf1 tapetum differentiation | Tapetum does not differentiate. | 4J, S2B | ||
| ii. pf2 tapetum/middle layer signaling | Tapetum is released from the degenerating middle layer. | 4L, S2D | ||
| iii. pf3, pf5 microspore shape | Microspores are irregular in shape. The two mutants have very similar phenotype. | 4N, S2F, S2G | ||
| iv. pf4 middle layer and tapetum degeneration function. | Tapetum and middle layer do not degenerate. | 4O, S2I |
The left-hand column lists the three major male-defective phenotypic classes and shows the total numbers of deletions found in each of them. The third column describes the general phenotype of each group (Roman numerals indicate subgroups).
The asexual class:
Anthers of both asx1 and asx2 mutants are similarly arrested before differentiation of the parietal layers and the mutants are thus completely sterile. These flowers have suppressed gynoecia, as in wild-type male flowers, and rudimentary anthers similar to those in developing flowers of wild-type female (see Farbos et al. 1999) (Figure 4A shows the wild-type female anther rudiments, and Figure 4B shows asx1).
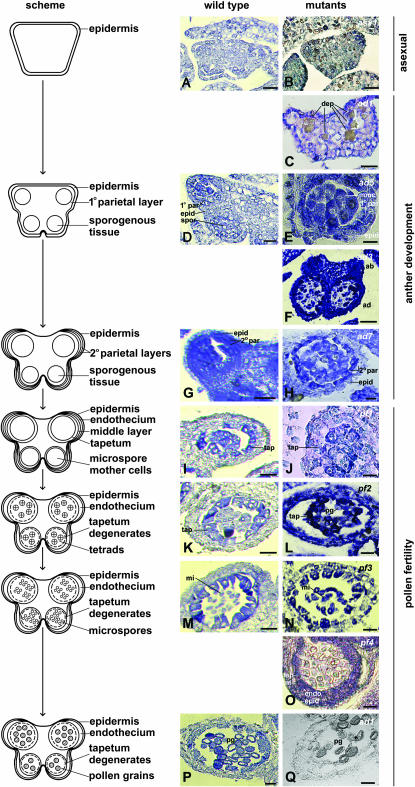
Figure 4.—
Histological analyses of S. latifolia wild type and Y chromosome deletion mutants. An outline of anther development is given on the left. The middle column shows sections of wild-type anthers, and the right-hand column displays sections of anther developmental mutants. Detailed descriptions of the wild-type anther development and Y-chromosome deletion mutant histology are given in supplemental Figures S1 and S2 at http://www.genetics.org/supplemental/. The anther rudiment in female plants stops developing before the formation of the parietal layers (A). The anthers of the Y chromosome deletion mutant asexua 1 (asx1) strongly resemble female anther rudiments (B). The inner subwhorl anthers of the anther development mutant ad1 also do not develop beyond this stage, probably because of the accumulation of deposits around the connective (C). Wild-type male anthers develop the primary parietal layer and the sporogenous tissue (D). In the anther development mutant ad6, the primary parietal layer is altered, but the sporogenous tissue is able to form microspore mother cells (E). In the ad3 mutant, abaxial locule development is arrested prematurely (F). Unlike the wild-type anthers, where no difference in the staining ability of the secondary parietal layers is observed (G), the inner secondary parietal layer of the ad7 mutant differentiates into a densely-stained tapetum-like layer (H). In the wild-type anthers, the tapetum becomes densely stained after the formation of all anther layers, and this process is connected with its differentiation (I). However, the tapetum is not stained in the pollen fertility mutant pf1 (J). During meiosis and tetrad formation, the tapetum starts to degenerate and its cells are released from each other but remain attached to the other anther layers (K). In the pf2 mutant, the tapetum is released from the anther layers and forms an agglutinate inside the anther (L). The wild-type microspores, when released from the tetrads, are round shaped (M), but in mutant pf3, the microspores are irregular in shape (N). Mutant pf4 forms round microspores, and the endothecial and tapetal cells do not degenerate, as deduced from the fact that the endothecium is present and the tapetal cells remain attached to each other (O). When compared to the wild type (P), both the anther locules and the pollen grains of the outer subwhorl of the mutant ad1 are abnormally small (Q). dep, Deposits; epid, epidermis; 1° par, primary parietal layer; spor, sporogenous cells; mmc, microspore mother cells; ab, abaxial locule; ad, adaxial locule; 2° par, secondary parietal layer; tap, tapetum; mi, microspores; pg; pollen grains. Bars, 10 μm (A–C, E, H, J, O), 25 μm (D, G, I, K–N, P, Q), and 50 μm (F).
The anther developmental class:
The seven mutants in this class (ad1–ad7) have anthers arrested at early stages during the formation of parietal and/or sporogenous layers and are therefore categorized as anther developmental mutants. The ad class includes deletions affecting different stamen whorls and distinct stamen parts, and different anther tissues are affected, and the timing of the defects also differs (Table 2). These mutants are again completely sterile either because they do not form pollen grains (ad2, ad4, ad6, and ad7) or because the few pollen grains produced are sterile (ad1, ad3, and ad5). These mutants can be further subcategorized into four distinct and stable phenotypic groups with specific defects, as follows:
i. The ad1 mutant has two types of defects. The inner, antipetalous subwhorl of anthers reaches only the four-lobed stage (i.e., the parietal layers are not yet formed, similarly to the asx mutants), such anthers accumulating deposits around the connective region (Figure 4C and supplemental Figure S2L at http://www.genetics.org/supplemental/). The outer, antisepalous anther subwhorl forms all anther layers found in wild-type plants (i.e., epidermis, endothecium, middle layers, and tapetum), but the pollen grains are abnormally small and nonviable (Figure 4Q and supplemental Figure S2K; for the wild type, see Figure 4P).
ii. In the ad6 mutant, developmental arrest occurs at the stage corresponding to the formation of parietal layers in the wild type (for the wild type, see the Figure 4D). Although only the primary parietal layer is formed, the microspore mother cells can undergo meiosis (Figure 4E and supplemental Figure S1E). The ad2 phenotype is very similar (supplemental Figure S1D).
iii. The ad3 mutant forms asymmetrically developed anthers; the adaxial locules are well developed (i.e., all anther layers, and microspores, form correctly), but the formation of abaxial locules is highly reduced (Figure 4F and supplemental Figure S1G). The ad5 mutant displays a similar asymmetry and phenotype (supplemental Figure S1H).
iv. In the ad7 mutant, the secondary parietal layer often appears to differentiate directly, without formation of all anther layers (Figure 4H and supplemental Figure S1K). The stage of developmental arrest corresponds to the formation of the secondary parietal layer in the wild type (Figure 4G). Mutant ad4 has a similar phenotype, except that anthers are highly variable while degenerating (supplemental Figure S1J).
The pollen fertility class:
In the “late” mutants with impaired pollen fertility (pf1–pf5), all anther parts are formed, but the microspores either degenerate after meiosis or else the pollen grains are sterile. These five independent pollen fertility mutants belong to four distinct phenotypic groups, each mutant having a defined tapetal defect, as follows (see also Table 2):
i. Mutant pf1 forms all anther layers (similarly to the wild type; Figure 4I), but the tapetum does not differentiate (Figure 4J and supplemental Figure S2B).
ii. Mutant pf2 forms all layers properly, like the wild type (for the wild type, see Figure 4K), but the tapetum is released from the degenerating middle layer (Figure 4L and supplemental Figure S2D).
iii. Unlike the wild type (Figure 4M), mutant pf3 microspores are irregular in shape (Figure 4N and supplemental Figure S2F). Mutant pf5 has similar defects (supplemental Figure S2G).
iv. In mutant pf4, all parts of the anther are properly formed, but later on, the middle layer and tapetum do not degenerate (Figure 4O and supplemental Figure S2I).
Location of Y regions with stamen indispensable functions:
To determine the number of male indispensable genes responsible for the mutant phenotypes, we ascertained their locations by arranging the Y chromosome deletions of each phenotypic group according to the presence or absence of molecular markers. The deletion results showed that male indispensable genes were located in only two or three Y regions. On the p arm, there were one or two regions containing the loci causing the asx and ad mutations, i.e., genes with mainly early stamen functions. All the pf loci (affecting late male functions) located to one region on the q arm.
The overlapping series of deletions at the distal end of the Y chromosome p arm (Figure 3) produced hermaphroditic (bsx) mutants. These deletions (up to 50% of the chromosome arm) indicate that the gynoecium suppressing factor, or female suppression sex determination gene of males (often denoted by GSF or by SuF), is located at this end of the chromosome (Lardon et al. 1999; also see Farbos et al. 1999), consistent with previous results (Westergaard 1958). The loci that, according to the current deletion map (Figure 3), are closest to the GSF region, correspond to mutants ad5–7 (which are codeleted with the ScDO5 marker), but the physical distance between the two classes of genes is unknown. ad-type defects are never seen in bsx mutants. The size range of the bsx deletions suggests that the distance between GSF and the closest ad loci must be >50% of the p arm. As explained above, the cytogenetic results indicate that the GSF-containing region of the Y does not contain genes controlling stamen development or pollen fertility functions. Consistent with the cytology, several markers that are never deleted in hermaphrodites have been shown to be codeleted in stamen function mutants (Figure 3). ScQ14 (Zhang et al. 1998) has thus been inferred to be close to a stamen-promoting factor or factors (Zluvova et al. 2005a) and SlY4 (Atanassov et al. 2001) to a male fertility factor or factors (Moore et al. 2003). Interestingly, plants with a range of male defects lack the ScQ14 marker, including all the asexual and anther-developmental mutants (asx and ad1–ad7) and also one of the pollen fertility mutants (pf4; see Figure 3 and further details below). In contrast, the deletions in the four mutants with only pollen fertility defects (pf1, pf2, pf3, and pf5), defining a locus or loci involved in late anther functions, lack the SlY4 gene, which is currently mapped on the q-arm. As all these deletions are cytologically small (Table 1 and Figure 3), these loci must be located close to SlY4 in a different region of the Y chromosome from the ScQ14 marker.
The asx and ad mutants thus probably have overlapping deletions around the ScQ14 marker, which may extend to different distances on either side (the physical sizes of the regions between ScQ14 and the adjacent markers on either side are unknown and could be large). The ad5 and ad6 mutants apparently also have a second, more distal, deleted region partially shared with the ad7 mutant (defined by the three consecutive markers MS4, ScD05, and SlY3; see Figure 3). Mutant ad5 lacks the first two markers and ad7 lacks the second two, while ad6 lacks only the marker believed to be in the middle, ScD05. Thus, the abnormal X–Y pairing, which is seen only in ad7, could be due to loss of a gene (or sequences) located in this distal region, since the deletion in this mutant appears to be more extensive on the side where SlY3 was mapped by Zluvova et al. (2005a). Alternatively, this gene or sequences could be located near the ScQ14 region, since ad7 may have a deletion that extends beyond the region deleted in the other ad mutants but not as far as SlY4 (since the pf mutants have normal chromosome pairing).
The mutants asx1 and asx2, in which, as just explained, ScQ14 is absent, have large deletions of the Y chromosome p arm (Table 1). These mutants lack the stamen-promoting, sex-determining function, often denoted SPF (Farbos et al. 1999). Since asx2 has a cytologically smaller deletion than asx1, the stamen-promoting, sex-determining gene must lie within the region corresponding to the asx2 deletion (24% of the p arm of the Y), and the distance between ScQ14 and the SPF locus (and a gene or genes essential for early stamen development, defined by the ad mutants) must thus be ≤24% of the p arm.
The probability of all the deletions with the ad phenotypes occurring in one region of the Y and all the pf deletions in another single region is very low, if each of the eight ad and pf groups (Table 2) is deleted for a different gene, assuming that the genes are randomly distributed on the Y chromosome. We discuss below possible explanations for the finding that there are apparently two distinct gene regions on the Y chromosome with indispensable roles in anther formation, one containing the asx and ad loci on the p arm (early stamen developmental factors) and one bearing the pf loci on the q arm (pollen fertility factors).
DISCUSSION
Our screen identified eight different phenotypic groups of Y-linked mutants with a broad range of stamen development and pollen fertility traits (Table 2) in addition to the sex determination functions corresponding to the asx and bsx mutations previously described. Defects in late stamen functions, notably tapetum malfunction, like those in our pf mutants have been reported repeatedly in various plant species (Vizir et al. 1994; Gorman and McCormick 1997; Sanders et al. 1999).
The asx and ad mutants are, however, unusual. In Arabidopsis thaliana, only a few mutants have been identified with defects in the specification, organization, or development of anther cell types, similar to those in our ad class (e.g., ad2 and ad4; see Vizir et al. 1994; Sanders et al. 1999). Mutants with such defects were also not found in large-scale screens for fertility defects using EMS or irradiation mutagenesis in other model species such as maize and tomato (Gorman and McCormick 1997; Neuffer et al. 1997). This suggests that the S. latifolia Y chromosome carries an unusual kind of locus with early stamen functions (or an unexpectedly large number of such loci) in addition to a locus that generates the more common type of mutations affecting late stamen functions.
Types of male-indispensable genes identified:
Since several types of mutations appeared more than once, the Y has probably been saturated with respect to mutations affecting indispensable male functions, i.e., all regions whose deletion can affect such functions have been identified. Our mapping of asx, ad, and pf classes shows that the corresponding deletions span fairly large Y chromosome regions (Table 2). The observed asx–ad phenotypes produced by overlapping deletions can thus be explained in two ways (similar reasoning can be applied to the pf deletions). One possibility is that the different phenotypes observed with deletions in a single region resulted from deletion of different loci, indicating that several different genes with male indispensable and closely related functions lie in this Y region. Alternatively, the region contains a major gene involved in early stamen development, plus other (modifier) genes that interact with this gene, so that the phenotype is different in each distinct deletion.
We cannot distinguish between these two possibilities. A variant of the second possibility above is that effects such as position effect variegation (PEV) could influence the expression of nondeleted genes located near the breakpoints. The fact that independent mutants within a group (e.g., the ad3 and ad5 mutants) were always phenotypically identical or very similar suggests that PEV is unlikely, since it generally leads to variable phenotypes with the same rearrangement (Weiler and Wakimoto 1998 and references therein).
Male specialization of the Y:
The screen identified almost exclusively male function genes, and the mutant phenotypes are dominant or at least semidominant. Dominant mutations are generally much less frequent than recessive ones (Orr 1991; for an estimate in S. latifolia, see Lardon et al. 1999), so that it is not expected that one will find many such mutations in a screen. For male reproductive functions, however, the theory outlined in the Introduction predicts that Y chromosomes will be enriched for such genes, due either to (i) evolution of Y-linked alleles that actively increase male functions relative to their X-linked homologs (so that loss of the Y copy impairs male function), or (ii) transposition of genes onto the Y but not to the X; in addition, loss of the SPF should lead to complete sterility, since the X chromosome has a nonfunctional allele at this locus (defined as the locus that is mutated in females and causes their male sterility). Y chromosome genes of either type i or ii might have deleterious effects in females and fit into the category of sexually antagonistic genes proposed by Rice (1987) that are expected to accumulate on the Y once dioecy evolves.
In S. latifolia, the SlAP3Y gene is already known to be an example of recruitment to the Y of an autosomal gene, whose function has subsequently become more male specialized (Matsunaga et al. 2003). This gene appears to map in the asx-ad region (Figure 3). Although it is not possible to tell which of the two categories of changes mentioned above has been involved in cases other than this, the results described above suggest that the S. latifolia Y chromosome has started to undergo the predicted changes and carries several genes promoting male reproductive functions that cannot be replaced by X-linked alleles or autosomal genes, i.e., has evolved some male specialization. The process may be accentuated by degeneration and subsequent deletion of Y-linked genes not involved in male function (such as the MROS3 gene; reviewed in Charlesworth and Charlesworth 2000), but there is so far little evidence for this in S. latifolia.
Early and late stamen functions define distinct Y regions:
According to the hypothesis proposed by Charlesworth and Charlesworth (1978), male-benefit genes should start to accumulate on the proto-Y chromosome while it still recombines with the proto-X, forming clusters around the initial sex-determining genes. Once a nonrecombining region has evolved, male-benefit genes will continue to evolve throughout the Y-specific region (Rice 1987).
Each of the phenotypic categories of Y-linked mutants described above (bsx, asx-ad, and pf) is associated with deletions of specific parts of the Y chromosome, and the three major categories of mutants are each found only in deletions of a single Y region. These results thus suggest only two distinct regions with Y-specific genes having roles in anther formation. One region, located on the Yp arm, contains the gene(s) responsible for all nine early stamen developmental mutants, including the asx mutants and the four ad groups defined in Table 2 (all nine of these mutants are deleted for the ScQ14 marker; see Figure 3). In contrast, four of the five late stamen mutants (pf) have deletions in a different region, on the q arm, defined by loss of the SlY4 marker. This distinct pattern would be highly unlikely if these mutants represent 14 genes scattered throughout the Y chromosome.
The female suppression region (containing the GSF gene defined by the bsx mutants) is large (about one-half of the distal Y chromosome p arm) but contains no genes indispensable for stamen functions. A stamen-promoting region is defined by the asx mutants. This region, including the genes indispensable for early stamen functions that are deleted in all 7 ad mutants and 1 of the 5 pf mutants, defines the proximal ∼20% of the p arm. It may include the gene that is mutated or absent in females that causes their male deficiency, which may correspond to the locus defined by our asx mutations and thus represents the Y chromosomal SPF gene. If the asx-ad region contains several different genes with male-indispensable functions (see above), any of them could play the role of a male sex determination function and could thus be the SPF gene.
Finally, very late functions involved in pollen fertility (defined by the pf mutants) are located distant from the sex determination regions in the current S. latifolia Y, close to the breakpoint of the large Y inversion inferred to lie between SlY1 and SlY4 (Figure 1; see also below). The inversion must have occurred recently, as genes with high and low X–Y sequence divergence (Filatov 2005b; Nicolas et al. 2005) are found in adjacent locations in the Y map (Zluvova et al. 2005a). If the recent ancestral state was the same gene order as the current X chromosome, the pf gene(s) must thus have been located either near the SlY1 gene, i.e., close to the PAR on the q arm or else at the end of the arm distant from the PAR (i.e., near the GSF locus on the p arm). On the second hypothesis, the inversion (Figures 1 and 3) may be reinforcing reduced X–Y recombination either directly through the well-known effect of inversions in causing lower survival of recombinant gametes or zygotes due to chromosome breaks, or as a secondary consequence of allowing sequence divergence of this region of the Y.
Meiotic X–Y pairing:
Three of our mutants have altered X–Y pairing in meiosis. Among them, the ad7 deletion clearly indicates that the Y chromosome carries elements or functions essential for X–Y pairing arrest. S. latifolia mutants with pairing along the sex chromosomes may have been observed by Westergaard (1946; type Y2, with the differential or p arm only about one-fourth as long as the pairing arm), but he interpreted his cytological observations in terms of a translocation of a part of the X chromosome to the Y, with loss of almost all of the Yp arm. This interpretation cannot explain the ad7 deletion, because this mutant's Y chromosome is only slightly smaller than the wild-type Y (Table 1) and still possesses most of the Y-specific markers we tested, including markers from most Y regions (Figure 3 and supplemental Table S2 at http://www.genetics.org/supplemental/). The cause of the lack of pairing throughout most of the X and Y chromosomes is unknown. A reinversion in the mutant of the putative inverted region of the Y chromosome (Figure 1) (Zluvova et al. 2005a) seems implausible. If this is correct, the ad7 mutation indicates that the Y carries gene(s) or sequences restricting X–Y pairing. It is not possible to test whether pairing in the ad7 deletion affects X–Y recombination, because the mutant is male sterile (but restoration of recombination seems unlikely given the large sequence divergence that is likely in this region of the Y).
The region deleted when the pairing function is mutated is not close to the pseudo-autosomal region (which might suggest that it was involved in loss of recombination in the most recently evolved stratum of the sex chromosomes). Interestingly, however, it is close to the region carrying the SlY3 gene (Figure 1), a gene pair with the highest level of X–Y sequence divergence yet observed (Nicolas et al. 2005; Bergero et al. 2007). As explained above, this region also contains a male-promoting gene, which could be one of the primary sex determination loci. In this region, the S. latifolia X and Y chromosomes are as highly diverged as homeologous chromosome pairs in many allopolyploid plant species. This lack of close homology may not necessarily prevent chromosome pairing. A classical example of homeologous chromosome pairing occurs in wheat, and it is suppressed by an allelic variant of the Ph1 locus (Riley and Chapman 1958; Martinez-Perez et al. 2001; Griffiths et al. 2006). It is possible that the S. latifolia genome has evolved a similar mechanism to restrict sex chromosome pairing.
Whatever the mechanism, our results are consistent with restricted X–Y recombination in S. latifolia having evolved in steps, as suggested by previous results on differences in sequence divergence of X–Y gene pairs spanning most X–Y regions (Filatov 2005a; Nicolas et al. 2005; Bergero et al. 2007). The meiotic behavior of the ad7 mutant supports this by indicating that pairing can be stopped through sex chromosome pair-specific processes. In S. latifolia, more genes (for estimating X–Y divergence, which can be used to infer when recombination ceased) and FISH experiments with BAC clones will in the future allow better inferences of the extent of X–Y rearrangements, including detecting other, smaller inversions that may have occurred at different times in the evolution of the Y chromosome (some of which might have been involved in suppressing its recombination with the X).
Acknowledgments
This research was supported by the Grant Agency of the Czech Republic (204/05/P505 to J.Z., 521/05/2076 to B.J., and 521/06/0056 to B.V.), by the Institutional Research Plan (AVOZ50040507), and through the bilateral French-Bulgarian program Rila06316YC. D.C.'s work was funded by the Natural Environment Research Council of the UK.
Note added in proof: The rearrangements of the genomic material on the Y chromosome are not confined to a single pericentric inversion, as shown by Hobza et al. (R. Hobza, E. Kejnovsky, B. Vyskot and A. Widmer, 2007, The role of chromosomal rearrangements in the evolution of Silene latifolia sex chromosomes. Mol. Genet. Genomics, in press). This does not affect any of our conclusions, except that it makes the hypothesis of a reinversion, which we reject in the discussion, even less plausible.
References
- Ainsworth, C., J. Parker and V. Buchanan-Wollaston, 1998. Sex determination in plants. Curr. Top. Dev. Biol. 38: 167–223. [DOI] [PubMed] [Google Scholar]
- Atanassov, I, C. Delichere, D. A. Filatov, D. Charlesworth, I. Negrutiu et al., 2001. Analysis and evolution of two functional Y-linked loci in a plant sex chromosome system. Mol. Biol. Evol. 18: 2162–2168. [DOI] [PubMed] [Google Scholar]
- Bergero, R., A. Forrest, E. Kamau and D. Charlesworth, 2007. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175: 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, R., 2000. Negative genetic correlation between male sexual attractiveness and survival. Nature 406: 67–70. [DOI] [PubMed] [Google Scholar]
- Bull, J. J., 1983. Evolution of Sex Determining Mechanisms. Benjamin/Cummings Publishing Company, Menlo Park, CA.
- Carvalho, A. B., 2002. Origin and evolution of the Drosophila Y chromosome. Curr. Opin. Genet. Dev. 12: 664–668. [DOI] [PubMed] [Google Scholar]
- Charchar, F. J., M. Svartman, N. El-Mogharbel, M. Ventura, P. Kirby et al., 2003. Complex events in the evolution of the human pseudoautosomal region 2 (PAR2). Genome Res. 13: 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1991. The evolution of sex chromosomes. Science 251: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., 2002. Plant sex determination and sex chromosomes. Heredity 88: 94–101. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., and D. Charlesworth, 1978. Model for evolution of dioecy and gynodioecy. Am. Nat. 112: 975–997. [Google Scholar]
- Charlesworth, B., and D. Charlesworth, 2000. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., B. Charlesworth and G. Marais, 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Desfeux, C, S. Maurice, J. P. Henry, B. Lejeune and P. H. Gouyon, 1996. Evolution of reproductive systems in the genus Silene. Proc. R. Soc. Lond. Ser. B 263: 409–414. [DOI] [PubMed] [Google Scholar]
- Donnison, I. S., J. Siroky, B. Vyskot, H. Saedler and S. R. Grant, 1996. Isolation of Y chromosome-specific sequences from Silene latifolia and mapping of male sex-determining genes using representational difference analysis. Genetics 144: 1893–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, P. J., and N. A. Affara, 2006. Spermatogenesis and sex chromosome gene content: an evolutionary perspective. Hum. Fertil. 9: 1–7. [DOI] [PubMed] [Google Scholar]
- Farbos, I., J. Veuskens, B. Vyskot, M. Oliveira, S. Hinnisdaels et al., 1999. Sexual dimorphism in white campion: deletion on the Y chromosome results in a floral asexual phenotype. Genetics 151: 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov, D. A., 2005. a Evolutionary history of Silene latifolia sex chromosomes revealed by genetics mapping of four genes. Genetics 170: 975–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov, D. A., 2005. b Substitution rates in a new Silene latifolia sex-linked gene, SlssX/Y. Mol. Biol. Evol. 22: 402–408. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A., 1931. The evolution of dominance. Biol. Rev. 6: 345–368. [Google Scholar]
- Gibson, J. R., A. K. Chippindale and W. R. Rice, 2002. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc. R. Soc. Lond. Ser. B 269: 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman, S. W., and S. McCormick, 1997. Male sterility in tomato. Crit. Rev. Plant Sci. 16: 31–35. [Google Scholar]
- Griffiths, S., R. Sharp, T. N. Foote, I. Bertin, M. Wanous et al., 2006. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752. [DOI] [PubMed] [Google Scholar]
- Hinnisdaels, S., A. Lardon, N. Barbacar and I. Negrutiu, 1997. A floral third whorl-specific marker gene in the dioecious species white campion is differentially expressed in mutants defective in stamen development. Plant Mol. Biol. 35: 1009–1014. [DOI] [PubMed] [Google Scholar]
- Hobza, R., P. Hrusakova, J. Safar, J. Bartos, B. Janousek et al., 2006. MK17, a specific marker closely linked to the gynoecium suppression region on the Y chromosome in Silene latifolia. Theor. Appl. Genet. 113: 280–287. [DOI] [PubMed] [Google Scholar]
- Iwase, M, Y. Satta, Y. Hirai, H. Hirai, H. Imai et al., 2003. The amelogenin loci span an ancient pseudoautosomal boundary in diverse mammalian species. Proc. Natl. Acad. Sci. USA 100: 5258–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent-First, M., 2000. The Y chromosome and its role in testis differentiation and spermatogenesis. Semin. Reprod. Med. 18: 67–80. [DOI] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1999. a Four evolutionary strata on the human X chromosome. Science 286: 964–967. [DOI] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1999. b Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat. Genet. 21: 429–433. [DOI] [PubMed] [Google Scholar]
- Lardon, A., S. Georgiev, A. Aghmir, G. Le Merrer and I. Negrutiu, 1999. Sexual dimorphism in white campion: complex control of carpel number is revealed by Y chromosome deletions. Genetics 151: 1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel-Hardenack, S., E. Hauser, T. F. Law, J. Schmid and S. R. Grant, 2002. Mapping of sex determination loci on the white campion (Silene latifolia) Y chromosome using amplified fragment length polymorphism. Genetics 160: 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack, J., L. A. Raubeson, L. Cui, J. V. Kuehl, M. H. Fourcade et al., 2005. Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one's way out of the Felsenstein zone. Mol. Biol. Evol. 22: 1948–1963. [DOI] [PubMed] [Google Scholar]
- Lengerova, M., R. C. Moore, S. R. Grant and B. Vyskot, 2003. The sex chromosomes of Silene latifolia revisited and revised. Genetics 165: 935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm, A., and F. Breden, 2002. Sex chromosomes and sexual selection in poeciliid fishes. Am. Nat. 160: S214–224. [DOI] [PubMed] [Google Scholar]
- Marais, G., and N. Galtier, 2003. Sex chromosomes: how X–Y recombination stops. Curr. Biol. 13: R641–643. [DOI] [PubMed] [Google Scholar]
- Markova, M., M. Lengerova, J. Zluvova, B. Janousek and B. Vyskot, 2006. Karyological analysis of an interspecific hybrid between the dioecious Silene latifolia and the hermaphroditic Silene viscosa. Genome 49: 373–379. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez, E., P. Shaw and G. Moore, 2001. The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature 411: 204–207. [DOI] [PubMed] [Google Scholar]
- Matsunaga, S., E. Isono, E. Kejnovsky, B. Vyskot, J. Dolezel et al., 2003. Duplicative transfer of a MADS box gene to a plant Y chromosome. Mol. Biol. Evol. 20: 1062–1069. [DOI] [PubMed] [Google Scholar]
- McAllister, B. F., and A. L. Evans, 2006. Increased nucleotide diversity with transient Y linkage in Drosophila americana. PLoS ONE 1: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R.C., O. Kozyreva, S. Lebel-Hardenack, J. Siroky, R. Hobza et al., 2003. Genetic and functional analysis of DD44, a sex-linked gene from the dioecious plant Silene latifolia, provides clues to early events in sex chromosome evolution. Genetics 163: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutiu, I., B. Vyskot, N. Barbacar, S. Georgiev and F. Moneger, 2001. Dioecious plants. A key to the early events of sex chromosome evolution. Plant Physiol. 127: 1418–1424. [PMC free article] [PubMed] [Google Scholar]
- Neuffer, M. G., E. H. Coe and S. R. Wessler, 1997. Mutants of Maize. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Nicolas, M., G. Marais, V. Hykelova, B. Janousek, V. Laporte et al., 2005. A gradual and ongoing process of recombination restriction in the evolutionary history of the sex chromosomes in dioecius plants. PLoS Biol. 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1991. A test of Fisher's theory of dominance. Proc. Natl. Acad. Sci. USA 88: 11413–11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, J., and A. Ashworth, 1999. Evolutionary rate of a gene affected by chromosomal position. Curr. Biol. 9: 987–989. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914. [DOI] [PubMed] [Google Scholar]
- Riley, R., and V. Chapman, 1958. Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182: 713–715. [Google Scholar]
- Ross, M. T., D. V. Grafham, A. J. Coffey, S. Scherer, K. McLay et al., 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, P. M., A. Q. Bui, K. Weterings, K. N. McIntire, Y.-C. Hsu et al., 1999. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322. [Google Scholar]
- Sandstedt, S. A., and P. K. Tucker, 2004. Evolutionary strata on the mouse X chromosome correspond to strata on the human X chromosome. Genome Res. 14: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti, I., and L. F. Delph, 2006. Selective trade-offs and sex-chromosome evolution in Silene latifolia. Evolution 60: 1793–1800. [PubMed] [Google Scholar]
- Scutt, C.P., M. Oliveira, P. M. Gilmartin and I. Negrutiu, 1999. Morphological and molecular analysis of a double-flowered mutant of the dioecious plant white campion showing both meristic and homeotic effects. Dev. Genet. 25: 267–279. [DOI] [PubMed] [Google Scholar]
- Siroky, J., M. R. Castiglione and B. Vyskot, 1998. DNA methylation patterns of Melandrium album chromosomes. Chromosome Res. 6: 441–446. [DOI] [PubMed] [Google Scholar]
- Skaletsky, H., T. Kuroda-Kawaguchi, P. J. Minx, H. S. Cordum, L. Hillier et al., 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–837. [DOI] [PubMed] [Google Scholar]
- Vicoso, B., and B. Charlesworth, 2006. Evolution on the X chromosome: unusual patterns and processes. Nat. Rev. Genet. 7: 645–653. [DOI] [PubMed] [Google Scholar]
- Vizir, I. Y., M. L. Anderson, Z. A. Wilson and B. J. Mulligan, 1994. Isolation of deficiencies in the Arabidopsis genome by {gamma}-irradiation of pollen. Genetics 137: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyskot, B., and R. Hobza, 2004. Gender in plants: sex chromosomes are emerging from the fog. Trends Genet. 20: 432–438. [DOI] [PubMed] [Google Scholar]
- Vyskot, B., J. Siroky, R. Hladilova, N. D. Belyaev and B. M. Turner, 1999. Euchromatic domains in plant chromosomes as revealed by H4 histone acetylation and early DNA replication. Genome 42: 343–350. [PubMed] [Google Scholar]
- Weiler, K. S., and B. T. Wakimoto, 1998. Chromosome rearrangements induce both variegated and reduced, uniform expression of heterochromatic genes in a developmental-specific manner. Genetics 149: 1451–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard, M., 1946. Aberrant Y chromosomes and sex expression in Melandrium album. Hereditas 32: 419–443. [DOI] [PubMed] [Google Scholar]
- Westergaard, M., 1958. The mechanism of sex determination in dioecious flowering plants. Adv. Genet. 9: 217–281. [DOI] [PubMed] [Google Scholar]
- Winge, O., 1927. The location of eighteen genes in Lebistes reticulatus. J. Genet. 18: 1–43. [Google Scholar]
- Zhang, Y. H., V. S. Di Stilio, F. Rehman, A. Avery, D. Mulcahy et al., 1998. Y chromosome specific markers and the evolution of dioecy in the genus Silene. Genome 41: 141–147. [Google Scholar]
- Zluvova, J., B. Janousek, I. Negrutiu and B. Vyskot, 2005. a Comparison of the X and Y chromosome organization in Silene latifolia. Genetics 170: 1431–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zluvova, J., M. Lengerova, M. Markova, R. Hobza, M. Nicolas et al., 2005. b The inter-specific hybrid Silene latifolia x S. viscosa reveals early events of sex chromosome evolution. Evol. Dev. 7: 327–336. [DOI] [PubMed] [Google Scholar]