Abstract
We describe the most comprehensive study to date on gene expression during mouse inner ear (IE) organogenesis. Samples were microdissected from mouse embryos at E9–E15 in half-day intervals, a period that spans all of IE organogenesis. These included separate dissections of all discernible IE substructures such as the cochlea, utricle, and saccule. All samples were analyzed on high density expression microarrays under strict statistical filters. Extensive confirmatory tests were performed, including RNA in situ hybridizations. More than 5000 genes significantly varied in expression according to developmental stage, tissue, or both and defined 28 distinct expression patterns. For example, upregulation of 315 genes provided a clear-cut “signature” of early events in IE specification. Additional, clear-cut, gene expression signatures marked specific structures such as the cochlea, utricle, or saccule throughout late IE development. Pathway analysis identified 53 signaling cascades enriched within the 28 patterns. Many novel pathways, not previously implicated in IE development, including β-adrenergic, amyloid, estrogen receptor, circadian rhythm, and immune system pathways, were identified. Finally, we identified positional candidate genes in 54 uncloned nonsyndromic human deafness intervals. This detailed analysis provides many new insights into the spatial and temporal genetic specification of this complex organ system.
MORE than 10% of the human population has hearing or balance disorders; two-thirds of these are between the ages of 21 and 65. One newborn out of 1000 suffers from profound deafness (Parving 1993; Mehl and Thompson 1998), and up to 15% of children between 6 and 19 years of age have some form of hearing loss (Marazita et al. 1993; Niskar et al. 1998). Environmental causes play a significant role in this, but genetic determinants are estimated to account for at least one-half of all congenital hearing and balance disorders. In light of these facts, it is clearly important to understand the genetic program of normal development for the mammalian inner ear (IE). One approach to that end is to screen for single gene defects that result in either balance or hearing deficiencies (i.e., abnormal development of the IE). This has been a productive route in the mouse where both auditory and balance phenotypes are relatively easy to score (Avraham 2003). However, such single gene approaches are slow to yield information on critical pathways or networks of genes. In this article we describe the most comprehensive analysis to date on transcriptional changes in the developing mammalian IE, with an emphasis on discovering the pathways and networks that underlie organogenesis in this complex set of structures.
The mature mammalian IE has two major components: the vestibular and auditory organs. The vestibular organ senses balance and changes in movement. It contains the three semicircular canals that sense angular acceleration and the utricle and saccule, both of which are responsible for sensing gravity and linear acceleration. The auditory organ consists of the coiled cochlea, which senses sound. Within both of these organs a specialized sensory epithelium converts mechanical actions into electrical potentials. These epithelia contain sensory hair cells—mechanoreceptors that initiate action potentials in response to sound or movement—as well as surrounding supporting cells. Damage to this small population of hair cells is a major cause of hearing loss. There are numerous other cell types in the IE that are also required for the mechanical, electrical, and structural aspects of hearing and balance. Examples of such cell types are the nonsensory supporting cells surrounding the hair cells (Raphael and Altschuler 2003), those of the stria vascularis on the lateral wall of the cochlear duct, responsible for the production of the endocochlear electrical potential (Takeuchi et al. 2000), and those of the various membranes on which the sensory organs rest and that separate the different compartments of the IE (Sulik 1995; Raphael and Altschuler 2003).
The morphological events that accompany organogenesis of the IE and some of the signaling molecules involved in the patterning of the IE, have been described in some detail (Sulik 1995; Gallagher et al. 1996; Morsli et al. 1998; Fritzsch et al. 1998; Cantos et al. 2000; Kelley et al. 2005). In the mouse, the IE first becomes evident as an otic placode at embryonic day (E) 8.5. These placodes are bilateral thickenings of the lateral ectoderm above the hindbrain. These invaginate and form otic cups/pits by E9 and eventually otic vesicles/otocysts by E9.5. The otocyst elongates and forms a dorsal vestibular pouch and a ventral cochlear pouch. At around E12.5, the utricle, saccule, and the three semicircular canals of the vestibular organ become visually discernible. The sensory hair cells in the vestibular organ appear at about E13, a day earlier than they do in the cochlea (Ruben 1967; Anniko 1983; Lumpkin et al. 2003). Full development of the IE continues postnatally; the mouse IE does not become fully mature until three weeks after birth, but by E15 all of the major structures and cell types are already present.
One step toward understanding how the IE develops and functions in its entirety is to catalog the time and place of expression of all genes expressed within this complex organ. Currently, there are several resources that list information about some of the protein-coding genes expressed in different regions of the IE and/or whether any are known to cause an IE defect when mutated (http://www.sanger.ac.uk/PostGenomics/mousemutants/deaf/; http://www.jax.org/hmr/map.html; http://webhost.ua.ac.be/hhh/; http://www.ihr.mrc.ac.uk/hereditary/genetable/index.shtml) (Robertson et al. 1994; Anagnostopoulos 2002; Resendes et al. 2002; Beisel et al. 2004; Kelley et al. 2005). These assist in identifying genes that function in the IE, but they fail to provide a dynamic temporal pattern of expression of such genes over a larger timescale. This is primarily due to the fact that most studies to date have sampled genes from just one particular time point and many have sampled genes from tissues that are quite heterogeneous. Hawkins et al. (2006) and others (Robertson et al. 1994; Resendes et al. 2002; Beisel et al. 2004) have constructed cDNA libraries from IE tissues, but these resources, while valuable, are not comprehensive. Several microarray expression profiling studies of the IE also exist (Chen and Corey 2002; Hawkins et al. 2003; Lin et al. 2003; Liu et al. 2004; Toyama et al. 2005). While these are undoubtedly useful in identifying genes expressed at particular stages of IE development, they are limited by the fact that they either only provide a static view of gene expression or describe expression of a selected category of genes at multiple stages that are separated from one another by large gaps. Moreover, such studies do not cover all the sensory regions of the IE.
Here, we describe a new resource for data mining and discovery of genes involved in IE organogenesis. This involved large-scale gene expression profiling across all stages and substructures of IE development and included the discovery of novel pathways and patterns that act during this complex process. Specifically, we describe 28 distinct patterns of gene expression on the basis of tissue type, developmental stage, or a combination of both. Genes from each type of pattern were used to identify 53 significant biological signaling pathways potentially active during IE development. Many of these pathways have not previously been implicated in IE organogenesis. We have also validated the expression of a selected number of genes using independent means such as RNA in situs and semiquantitative RT–PCR. Finally, we present a large number of new candidate genes that map to uncloned human deafness intervals. Our entire data set is freely available online [Gene Expression Omnibus (GEO) series accession no. GSE7536] and should provide a valuable source of new individual genes and networks for further genetic investigations.
MATERIALS AND METHODS
IE dissections:
Timed pregnant CBA/J mice were euthanized with carbon dioxide, and IE tissues were dissected as described (Lumpkin et al. 2003). For each gestational stage, two biological replicates were collected, i.e., two pools of tissues from different identical staged litters. From E9–E10, IE epithelia from five to eight embryos were pooled. From E10.5 to E12, the ventral cochlear region and the dorsal vestibular region (without the endolymphatic duct) were separated and pooled separately for each stage from five to eight embryos. For stages E12.5–E15, the cochleae and the saccules from three to six embryos were separately pooled, whereas the utricles and the ampullae of the three semicircular canals were combined and pooled together (without the endolymphatic duct and canals). This utricle/ampullae mixture is referred to as “utricles” in the text. The noninner ear (NIE) tissues were also obtained from each stage and pooled as follows: stage E9 NIE tissues were pooled from four to five embryos per replicate; from E9.5 to E10.5 NIE was pooled from two to seven embryos; for E11–E15 NIE tissue was pooled from two to four embryos. Thus, a total of 29 IE and 3 NIE samples were obtained, each in duplicate, from 13 distinct developmental stages.
RNA isolation, cDNA synthesis, target synthesis:
Total RNA was isolated and processed as described (Hawkins et al. 2003). Total RNA was resuspended in either 7–10 μl (for stages E9–E10.5) or 15–20 μl (for stages E11–E15) of H2O. RNA quality was assessed by agarose gel electrophoresis of an aliquot of total RNA. PolyA RNA was isolated and converted to cDNA as previously described (Hawkins et al. 2003). This cDNA was then PCR amplified for a total of 12 cycles. Biotin-labeled target (cRNA) was derived from this cDNA by in vitro transcription reactions using the BioArray HighYield RNA transcript labeling kit (ENZO Life Sciences, New York) and a T7 promoter embedded within the 3′ end of the cDNA PCR products. Labeled cRNA was purified and eluted in water using an RNA purification kit (QIAGEN, Valencia, CA) following the manufacturer's instructions.
Array hybridization and analysis of differential expression:
A total of 20 μg of cRNA were fragmented, hybridized to MOE430A_2 Affymetrix arrays, and scanned following standard Affymetrix protocols. Supplemental Materials and Methods (http://www.genetics.org/supplemental/) extensively describes all aspects of data normalization, intensity filtering, and the generation of lists of probe sets/genes with specific expression patterns [e.g., early–middle–late (EML) analysis, etc.].
Gene ontology annotations:
Genes from various “present” lists and expression pattern types were uploaded in eGoN (http://www.genetools.microarray.ntnu.no/egon/index.php), a web-based tool for classifying multiple gene lists simultaneously on the basis of gene ontology (GO) annotations and finding statistically over-represented categories (cumulative hypergeometric probability of ≤0.05). All gene lists were uploaded using Affymetrix probe sets (only one per unique gene), and tests were carried out using the “Master-Target” option.
Identifying significant biological pathways:
The various lists of differentially expressed genes were analyzed by Ingenuity pathways analysis (IPA) (Ingenuity Systems, Redwood City, CA). Genes from each of the individual 28 expression patterns together with their ratios (≥1.5-fold) were uploaded in IPA using Entrez IDs as gene identifiers to identify significant biological pathways. Genes that did not have Entrez IDs in the Affymetrix NetAffx database were instead represented by probe set IDs. All genes within the resulting networks (focus and nonfocus genes) were exported from IPA. We next determined whether the nonfocus genes from each list were “present” or “absent” in our data set regardless of whether or not they were differentially expressed. These expanded lists were then re-uploaded in IPA to determine pathway significance. Note that the ratio of all nonfocus genes was designated as negative three. Only pathways that had at least two genes differentially expressed were considered. For the “middle” and “late” analyses, we re-uploaded focus genes combined with nonfocus genes that were both present and at the same time passed the ANOVA test P-value cutoff of ≤0.005.
Whole mount RNA in situ hybridizations:
PCR products were amplified (with primers that contained a T7 promoter at one or the other end) using cDNA from various developmental stages throughout the time course. The following are the specific nucleotides amplified: FoxP1 nucleotides 1341–1546, NM_053202; Hey2 nucleotides 1333–1437, NM_013904; Irx5 nucleotides 1600–1854, NM_018826; and Clu nucleotides 1291–1540, NM_013492. These were sequence verified and used for in vitro synthesis of DIG-labeled RNAs using Ambion's T7 megascript RNA synthesis kit. See supplemental Materials and Methods for sequences of the probes. Approximately 1 ng/μl of the labeled RNA was used in in situ hybridizations that were carried out as described (http://axon.med.harvard.edu/∼cepko/protocol/ctlab/ish.ct.htm). Hybridization was carried out at 58–60°. All steps were carried out either on whole IEs still in temporal bone (stages E13 and beyond) or on whole embryos (E11.5 and younger). After signal developed, whole IEs were dissected from the embryos E11.5 and younger, and tissues from all stages were incubated in 3–5 mg/ml dispase (Gibco, Grand Island, NY) at 37° for 1–2 hr. The IE epithelium was then dissected free of the cartilage and other NIE tissue and photographed.
RESULTS
Microdissection of IE and adjacent NIE tissues:
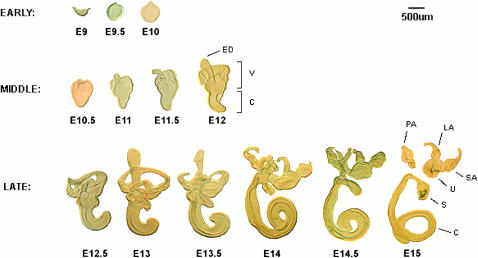
In all of the analyses described in this study we employed the Affymetrix mouse MOE430A_2 gene chip. This gene chip contains 14,065 unique genes represented by a total of 22,626 individual probe sets. We microdissected IE structures at half-day intervals from E9, at a time when the IE is an otic cup of ∼500 μm diameter, up to E15 when all of the major structures of the IE are anatomically distinguishable, and also when the differentiation of hair and supporting cells has been well initiated in all of the six sensory organs (Ruben 1967; Anniko 1983; Lumpkin et al. 2003). A total of 13 IE developmental stages were collected at half-day intervals. Figure 1 shows examples of microdissected structures used in this study and illustrates the attention that was paid to obtaining high quality samples. Tissues from E9 to E10, classified as “early,” were gene expression profiled in their entirety. Those from E10.5 to E12, designated as “middle” stages, were separated into the dorsal vestibular organ and the ventral cochlea. Each of these was then separately analyzed on gene chips. Tissues from “late” stages, i.e., from E12.5 to E15, were separated into three parts: the cochlea, the saccule, and the utricle (the latter being combined with the superior, posterior, and lateral ampullae). These three tissue types were then separately profiled. Thus, a total of 29 IE samples were analyzed from the 13 developmental stages, each being collected in duplicate (from different mouse litters). In addition to these tissues, we also dissected adjacent noninner tissues (NIE) from areas in close proximity to the IE tissue at each stage. This enabled us to subsequently estimate whether observed changes in gene expression were specific to the IE or a more broad reflection of stage-specific changes across many cell types. Specifically, NIE tissue from E9, consisting primarily of a mixture of neuroepithelial and mesenchymal cells, was profiled by itself. NIE tissues from E9.5 to E10.5, consisting mostly of ganglia, mesenchymal, and vascular cells, were combined and profiled together. Finally, NIE tissues from E11 to E15, mostly composed of mesenchyme, ganglia, vascular cells, the modiolus, and early cartilage were pooled and profiled together.
Figure 1.—
Representative microdissected IE tissues from mouse developmental stages E9 to E15 that were used for expression profiling. The top dorsal region is the vestibular organ, and the bottom ventral region is the cochlea. Early, middle, and late refer to the categories into which the structures were classified for data analysis (see Table 1). Early tissues were profiled whole, whereas the vestibular organ (V) and cochlea (C) from middle were profiled separately. In the late category, the cochlea and the saccule (S) were profiled individually. The utricle (U), posterior ampulla (PA), lateral ampulla (LA), and the superior ampulla (SA) were pooled and profiled together. The endolymphatic sac (ES) and the three semicircular canals were not profiled.
Measures of reproducibility and reliability:
In all microarray studies, and particularly those performed with microdissected samples that may vary in quality, it is important to determine the limits of reliability and reproducibility of such a large data set. In this regard, we performed four types of independent tests on our profiling data to check these parameters, in addition to the confirmatory RNA in situs described below [and others in supplemental materials (http://www.genetics.org/supplemental/)]. These tests are described in detail in supplemental Materials and Methods and in all cases provided strong confirmation of the data quality. Analysis of genes scored as present or absent, regardless of differential expression, is also provided in supplemental Materials and Methods.
Identifying classes of differentially expressed genes:
Initially, we searched for genes whose expression exhibited a dramatic peak or valley in one sample (i.e., only in one tissue at one stage) relative to all others. We anticipated that some of these genes might represent transiently expressed effectors of developmental choices. A relatively small number of genes (109 in total) met this particular criterion. Of these, 22 were detectable (present) only in the sample where expression was upregulated and were not detectable (absent) in all other samples. A total of 18 were detectable in all samples except the one where expression was downregulated (see supplemental Table 7 at http://www.genetics.org/supplemental/ for a listing of all 109 genes). The expression patterns of the 109 genes across the entire developmental time course are shown in the form of self-organizing maps (SOMs) in Figure 2 (and in higher resolution as supplemental Figure 6). These SOMs represent a form of unsupervised clustering that group genes with similar patterns of expression across the time course (Golub et al. 1999; Tamayo et al. 1999; Reich et al. 2004). The centroids of Figure 2 have been arranged into five groups (A–E) according to the similarity of their gene expression patterns. Thus, all of the centroids in group A show genes (a total of nine, listed to the right of the centroids) that decrease in gene expression in NIE samples relative to the IE samples. This pattern of dramatic changes in the NIE relative to IE is the predominant one observed in this analysis. Groups B and C show patterns in which expression decreases (B) or increases (C) in the NIE. Many of the upregulated genes in the NIE are components of the cytoskeleton and/or the extracellular matrix such as collagens, glycans, and proteases (supplemental Table 7). In some cases, genes that change in expression in the NIE samples also show relatively large changes in expression in at least one IE sample. For example, the otoconin-90 gene, which encodes the major protein component of the otoconia (Verpy et al. 1999), is downregulated in the E9 placode and in the NIE tissues relative to all other samples (group B, centroid 0 in Figure 2). Within the IE, it appears to be detectably expressed in all samples except the placode at E9. Eight additional genes in centroids 13 and 14 of group B also exhibit this pattern of gene expression. Overall, a total of 90 genes show the predominant NIE pattern of up- or downregulation relative to the IE samples. The remaining 19 genes fall into two classes; 14 show upregulation in the E9 otic cup and also some increased level of expression in the NIE tissues. Examples include the Slc2a3 gene that encodes a solute carrier transporter and Hapln1, which encodes a hyaluronan and proteoglycan link protein. The final seven genes exhibit just one pattern of expression; upregulation of expression in the E15 cochlea (group E of Figure 2). This group includes the insulin-like growth factor-1 gene (Igf1), which is required for the normal post-natal survival, maturation, and differentiation of the cochlear ganglion cells, and also for the normal innervation of cochlear sensory hair cells (Camarero et al. 2001). The significance of a spike in expression at E15, however, remains to be investigated further. Also in Figure 2E is the gene encoding a meteorin-like protein (Metrnl), which may play a role in axonal guidance and network formation (Nishino et al. 2004), the Calb1 gene that encodes the calcium-binding protein calbindin-28K (Dechesne and Thomasset, 1988), the gene-encoding integrin α-8 [which, when knocked out in the mouse, results in hair cells with malformed stereocilia (Evans and Muller, 2000)], two genes encoding proteoglycans (Spock1 and Dspg3), and the product of the mouse Silver locus (Silv). Expression of this latter gene is believed to be melanocyte specific (Theos et al. 2006). Its detection in the E15 cochlea may reflect the activity of the population of melanocytes in the developing stria vascularis.
Figure 2.—
Self-organizing maps (SOM) depicting the patterns of genes whose expression showed a peak or a valley in only one sample relative to all others. The y-axis is the expression level of a sample as a fraction of the average expression in all 32 samples. Fractions less than zero were converted to negative reciprocals. The order of the data points on the x-axis from left to right is: E9, E9.5, E10, cochleae from E10.5 to E12, vestibular organs from E10.5 to E12, cochleae from E12.5 to E15, utricles from E12.5 to E15, saccules from E12.5 to E15, and NIE tissues from E9, E9.5–E10.5, and E11–E15. The centroid ID (beginning with c and ending with a colon) and the total number of genes with that particular centroid pattern are indicated above each square (centroid). The dark blue line traces the average expression of all genes within each centroid. The top and bottom red lines trace the expression pattern on the basis of maximal and minimal expression values for each data point, respectively. Note that the maximal and minimal values, from left to right, are not necessarily from the same probe set. (A) Genes downregulated in a NIE tissue sample relative to all IE tissue samples. (B) Genes downregulated in a NIE tissue sample as well as one IE tissue sample. (C) Genes upregulated in a NIE tissue sample relative to all IE tissue samples. (D) Genes upregulated in a NIE tissue sample as well as one IE tissue sample. (E) Genes upregulated in the cochlea at E15. These maps are shown in higher resolution in supplemental Figure 6.
To detect broader trends in gene expression, rather than the more infrequent, discrete, and dramatic changes in expression described above, we conducted a series of comparisons between time points and tissues. These are shown in Tables 1 and 2 and fall into four types of analysis, which we named according to the types of gene expression pattern that they highlight: early–middle–late, middle, late, and IE vs. NIE. Within these analyses we then derived different patterns of gene expression. Details on each type of comparative analysis and the patterns of gene expression that they reveal are described below.
TABLE 1.
Classification of samples for data analysis
| Category | Samples in category | Subcategory | Samples in subcategory |
|---|---|---|---|
| Early (E) | E9–E10 | E9–E10 | E9–E10 |
| Middle (M) | E10.5–E12 | Based on stage | E10.5 |
| E11 | |||
| E11.5 | |||
| E12 | |||
| Based on tissue | Cochlea (Coch) | ||
| Vestibular organ (V) | |||
| Late (L) | E12.5–E15 | Based on stage | E12.5 |
| E13 | |||
| E13.5 | |||
| E14 | |||
| E14.5 | |||
| E15 | |||
| Based on tissue | Cochlea (C) | ||
| Utricle (U) | |||
| Saccule (S) |
Samples were assigned to three categories (early, middle, and late) on the basis of how many individual IE substructures at a particular developmental stage could be distinguished and separated from one another. Each category was then divided into subcategories on the basis of tissue type and developmental stage. Category E had only one subcategory consisting of arrays from E9 to E10. The M category had six subcategories (four based on stage and two based on tissue type). The L category had nine (six based on stage and three on tissue type).
TABLE 2.
Description of the 28 expression patterns identified in the data set
| Pattern description | Pattern abbreviation | Analysis |
|---|---|---|
| Genes upregulated in E relative to all subcategories of M and L | E | EML |
| Genes upregulated in at least one subcategory of M relative to E and relative to at least one subcategory of L | M | EML |
| Genes upregulated in at least one subcategory of L relative to E and relative to at least one subcategory of M | L | EML |
| Genes upregulated in E and in at least one subcategory of M relative to at least one subcategory of L | EM | EML |
| Genes upregulated in E and in at least one subcategory of L relative to at least one subcategory of M | EL | EML |
| Genes upregulated in at least one subcategory of M and in at least one subcategory of L relative to E | ML | EML |
| Genes upregulated in all cochlear relative to all vestibular samples | Cocha | Middle |
| Genes upregulated in all vestibular relative to all cochlea samples | Va | Middle |
| Genes upregulated in E10.5 and E11 relative to E11.5 and E12 | Yng-Mb | Middle |
| Genes upregulated in E11.5 and E12 relative to E10.5 and E11 | Old-Mb | Middle |
| Genes upregulated in E10.5 relative to all other stages in category M | E10.5-Upb | Middle |
| Genes downregulated in E10.5 relative to all other stages in category M | E10.5-Downb | Middle |
| Genes upregulated in E11 relative to all other stages in category M | E11-Upb | Middle |
| Genes downregulated in E11 relative to all other stages in category M | E11-Downb | Middle |
| Genes whose expression changed simultaneously on the basis of tissue type and developmental stage | Both-M | Middle |
| Genes upregulated in all cochlear relative to all utricular and saccular samples | Ca | Late |
| Genes upregulated in all utricular relative to all cochlea and saccular samples | Ua | Late |
| Genes upregulated in all saccular relative to all cochlear and utricular samples | Sa | Late |
| Genes upregulated in all cochlear and saccular samples relative to all utricular samples | CSa | Late |
| Genes upregulated in all cochlear and utricular samples relative to all saccular samples | CUa | Late |
| Genes upregulated in all utricular and saccular samples relative to all cochlear samples | USa | Late |
| Genes upregulated from E12.5 to E13.5 relative to E14 to E15 | Yng-Lb | Late |
| Genes upregulated from E14 to E15 relative to E12.5 to E13.5 | Old-Lb | Late |
| Genes upregulated in E14 relative to all other stages in category L | E14-Upb | Late |
| Genes downregulated in E14 relative to all other stages in category L | E14-Downb | Late |
| Genes whose expression changed simultaneously on the basis of tissue type and developmental stage | Both-L | Late |
| Genes upregulated in IE relative to NIE by twofold or more | IE-Up | IE vs. NIE |
| Genes downregulated in IE relative to IE by twofold or more | IE-Down | IE vs. NIE |
The 28 patterns of expression obtained from four types of analyses are described and the abbreviation for each is listed. EML, early–middle–late; IE, inner ear; NIE, noninner ear.
Patterns based only on tissue type.
Patterns based only on developmental stage.
Table 1 shows the groupings of stages and tissues that we employed in this study. All samples from E9 to E10 (three samples in total) were considered the early (E) category, while the ones from E10.5 to E12 were considered parts of the middle (M) category (these were further divided into four subcategories on the basis of developmental stage and two subcategories on the basis of tissue type). Samples from E12.5 to E15 were designated as late (L) and comprised six subcategories on the basis of developmental stage and three on the basis of tissue type. All of these various groups were compared to one another to identify genes that changed in expression only according to tissue type, or to developmental stage, or according to both tissue and time point. Table 2 describes the 28 different patterns of gene expression that were identified in these analyses, and Table 3 lists the number of genes that exhibited a >1.5-fold change in expression and also those that changed by >2-fold in each of the identified patterns. The lower fold-change cutoff was chosen so as to include genes known to be differentially expressed during these developmental stages and that also cause IE defects in mouse when mutated (e.g., Ctnnb1, Eya1, Eya4, Gja1, Gjb6, Notch1, and Sox10 among others).
TABLE 3.
Number of genes within each of the 28 patterns of expression
| No. of genes differentially expressed
|
||
|---|---|---|
| Pattern | By ≥1.5-fold | By ≥2-fold |
| E | 436 | 315 |
| M | 177 | 95 |
| L | 937 | 657 |
| EM | 1799 | 1328 |
| EL | 1033 | 633 |
| ML | 841 | 634 |
| Coch | 59 | 34 |
| V | 824 | 225 |
| Yng-M | 195 | 75 |
| Old-M | 218 | 82 |
| E10.5-Up | 71 | 33 |
| E10.5-Down | 202 | 80 |
| E11-UP | 57 | 17 |
| E11-Down | 182 | 91 |
| Both-M | — | 420 |
| C | 106 | 60 |
| U | 188 | 115 |
| S | 241 | 109 |
| CS | 43 | 31 |
| CU | 215 | 65 |
| US | 211 | 97 |
| Yng-L | 142 | 34 |
| Old-L | 188 | 84 |
| E14-Up | 171 | 43 |
| E14-Down | 499 | 151 |
| Both-L | — | 720 |
| IE-Up | — | 1410a |
| — | — | 384b |
| IE-Down | — | 226a |
| — | — | 66b |
The number of genes that change by ≥1.5-fold or ≥2-fold (with an estimated FDR of at most 0.5%) are shown for each type of pattern according to the abbreviations in Table 2. In EML analysis, the lowest boundary of fold changes was 1.5-fold after taking error into account. In M and L analyses, the number of genes indicated are those that were able to meet the two-way ANOVA P-value cutoff of 0.005 (see text and supplemental Materials and Methods for details). These had a lowest boundary of fold changes of 1.4-fold after taking error into account. Genes within expression patterns Both-M and Both-L do not have an expression ratio associated with them.
Genes that were either up- or downregulated in at least one-tenth of all IE samples relative to at least one NIE sample.
Genes that were either up- or downregulated in at least one-third of all IE samples relative to at least one NIE sample.
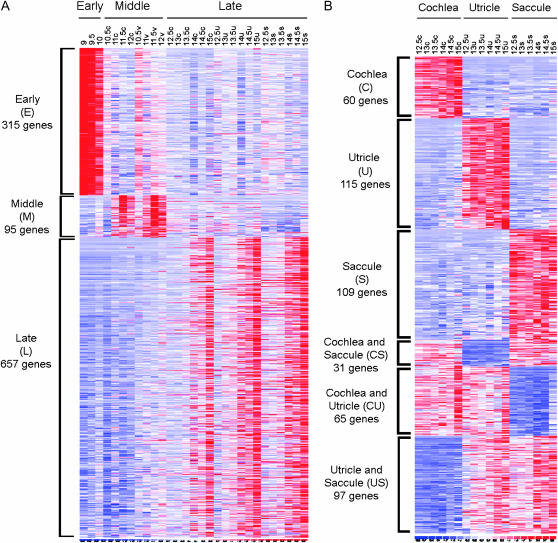
In EML analysis, we identified six different expression patterns. To accomplish this we compared the genes present in category E with those present in each subcategory of M and L. Significant analysis of microarrays (SAM) (Tusher et al. 2001) was used to identify those differentially expressed by at least 1.5-fold with an estimated false discovery rate (FDR) of ≤0.5%. On the basis of these comparisons, we then inferred the comparisons between each subcategory of M and L (see supplemental Materials and Methods). Genes that met the fold-change and FDR cutoffs in all the various comparisons were assigned to one of the expression patterns. Thus, for example, the EL pattern of expression contains genes that are upregulated in category E and also in at least one subcategory of L relative to M. The precision with which this analysis allowed us to group expression patterns is illustrated in Figure 3. This figure shows heat maps of genes that exhibit a twofold change or more in gene expression between various time point and tissue comparisons (red, upregulation; blue, downregulation). For example, Figure 3A shows the expression levels of genes that fall into the E, M, and L categories of gene expression. It is clear that the 315 genes in the E class are much more highly expressed in the E9 through E10 stages than at other stages and that the 95 middle stage genes are more highly expressed within the E10.5 through E12 stages than at other stages. These types of clear-cut differences can also be seen in Figure 3B where gene expression in later stages was broken down into tissue-specific classes. Thus, a set of 60 genes are clearly upregulated in cochlea, whereas 115 genes are more highly expressed in the utricle relative to the cochlea and saccule at E12.5–E15. The heat maps shown in Figure 3 (and additional ones that illustrate other comparative analyses) are available at higher resolution as supplemental Figure 3 (http://www.genetics.org/supplemental/). Table 4 lists examples of genes that exhibit large fold changes (threefold or more) in expression in only the cochleae (C), utricles (U), and saccules (S) shown in Figure 3B. Note that in the interests of space this is a partial listing. The complete list of these genes is presented in supplemental Table 4. Taken together, these sets of differentially expressed genes in various subgroupings provide discernible (and clear-cut) gene expression signatures for tissue type and/or time points within mouse IE development. For example, among the genes that appear to specify the cochlear signature are Tachykinin1, Clusterin, Gata3, Irx3, Irx5, FoxG1, Wnt7a, and Hey2 (among others). Among the utricle signature genes are Dlx1, Bmp2, Tbx3, Bmp6, Hes5, and FoxP1. Examples of saccule signature genes are Tachykinin receptor-3, Lhx1, Vav3, Zic1, Rarb, and FoxD1. Some of the genes listed in Table 4 have been previously studied in specific areas of the IE (e.g., Gata3), but these lists provide many more additional candidates and clues to unraveling the developmental programming of each of these IE organs.
Figure 3.—
Gene expression heat maps that illustrate temporal and tissue-specific signatures. In both A and B, each column is an individual IE sample (indicated on top), and each row represents a gene. Expression level increases from blue to white to red. The number of genes indicated in each expression pattern denotes those whose expression peaked by at least twofold in that pattern with an estimated false discovery rate (FDR) of <0.5%. Refer to supplemental Table 4 for a listing of the individual genes and fold changes within these expression patterns. (A) Heat maps of genes exhibiting three of the six expression patterns resulting from EML analysis (Table 2). Specifically, 315 genes were highly expressed in early stages, 95 in middle stages, and 657 in late stages. (B) Heat maps of genes with six expression patterns based on tissue type alone resulting from L analysis (Table 2). Shown are 60 genes whose expression peaked only in the cochlea, 115 only in the utricle, 109 only in the saccule, 31 in cochlea and saccule, 65 in cochlea and utricle, and 97 in utricle and saccule. c, cochlea; v, vestibular organ; u, utricle; s, saccule.
TABLE 4.
Examples of signature genes
| Gene description | Symbol | Entrez | Fold change (and pattern) |
|---|---|---|---|
| Carbonic anhydrase 13 | Car13 | 71934 | 3.3 (C) |
| Carboxylesterase 3 | Ces3 | 104158 | 11.2 (C) |
| Clusterin | Clu | 12759 | 17.1 (C) |
| Cytochrome P450, family 26, subfamily a, polypeptide 1 | Cyp26A1 | 13082 | 18.1 (C) |
| Endothelin converting enzyme-like 1 | Ecel1 | 13599 | 3.3 (C) |
| ES cell derived homeobox | Ehox | 194856 | 5.0 (C) |
| Forkhead box G1 | FoxG1 | 15228 | 6.3 (C) |
| Follistatin | Fst | 14313 | 26.2 (C) |
| GATA binding protein 3 | Gata3 | 14462 | 13.1 (C) |
| Gap junction membrane channel protein β 2 | Gjb2 | 14619 | 5.9 (C) |
| Hairy/enhancer of split related with YRPW motif 2 | Hey2 | 15214 | 3.0 (C) |
| High mobility group AT-hook 2 | Hmga2 | 15364 | 5.1 (C) |
| Iroquois related homeobox 3 (Drosophila) | Irx3 | 16373 | 9.5 (C) |
| Iroquois related homeobox 5 (Drosophila) | Irx5 | 54352 | 6.7 (C) |
| Solute carrier family 29 (nucleoside transporters), member 1 | Slc29A1 | 63959 | 4.0 (C) |
| Solute carrier family 39 (metal ion transporter), member 8 | Slc39A8 | 67547 | 9.1 (C) |
| Solute carrier family 7 (cationic amino acid transporter, y+ system), member 10 | Slc7A10 | 53896 | 4.4 (C) |
| Tachykinin 1 | Tac1 | 21333 | 36.1 (C) |
| Vascular endothelial growth factor C | Vegfc | 22341 | 5.6 (C) |
| Wingless-related MMTV integration site 7A | Wnt7a | 22421 | 3.1 (C) |
| Apolipoprotein B editing complex 2 | Apobec2 | 11811 | 23.4 (U) |
| BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) | Bambi | 68010 | 6.4 (U) |
| Bone morphogenetic protein 2 | Bmp2 | 12156 | 7.6 (U) |
| Bone morphogenetic protein 6 | Bmp6 | 12161 | 4.7 (U) |
| Calcium channel, voltage-dependent, α-2, δ-subunit 3 | Cacna2D3 | 12294 | 6.5 (U) |
| Cyclin D2 | Ccnd2 | 12444 | 3.1 (U) |
| CEA-related cell adhesion molecule 10 | Ceacam10 | 26366 | 3.6 (U) |
| Claudin 11 | Cldn11 | 18417 | 3.2 (U) |
| Claudin 8 | Cldn8 | 54420 | 8.1 (U) |
| Procollagen, type XIV, α 1 | Col14A1 | 12818 | 3.6 (U) |
| Distal-less homeobox 1 (Dlx1) | Dlx1 | 13390 | 27.5 (U) |
| Forkhead box P1 | FoxP1 | 108655 | 3.1 (U) |
| Glutamate receptor, ionotropic, AMPA2 (α-2) | Gria2 | 14800 | 17.8 (U) |
| Glutamate receptor, ionotropic, kainate 1 | Grik1 | 14805 | 3.5 (U) |
| Hairy and enhancer of split 5 (Drosophila) | Hes5 | 15208 | 8.7 (U) |
| Homer homolog 2 (Drosophila) | Homer2 | 26557 | 4.3 (U) |
| Potassium voltage-gated channel, Shal-related family, member 2 | Kcnd2 | 16508 | 22.9 (U) |
| Protocadherin 20 | Pcdh20 | 219257 | 3.5 (U) |
| Parathyroid hormone-like peptide | Pthlh | 19227 | 18.8 (U) |
| T-box 3 | Tbx3 | 21386 | 7.0 (U) |
| Carbonic anyhydrase 12 | Car12 | 76459 | 6.6 (S) |
| Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | Cdkn2B | 12579 | 5.7 (S) |
| Procollagen, type III, α-1 | Col3A1 | 12825 | 3.0 (S) |
| Chemokine (C-X-C motif) ligand 12 | Cxcl12 | 20315 | 4.3 (S) |
| Forkhead box D1 | FoxD1 | 15229 | 3.6 (S) |
| LIM homeobox protein 1 | Lhx1 | 16869 | 6.3 (S) |
| Mesenchyme homeobox 2 | Meox2 | 17286 | 3.7 (S) |
| Membrane metallo endopeptidase | Mme | 17380 | 3.4 (S) |
| Myosin 1H | Myo1H | 231646 | 6.5 (S) |
| Nidogen 1 | Nid1 | 18073 | 3.1 (S) |
| Otoancorin | Otoa | 246190 | 9.0 (S) |
| PDZ domain containing RING finger 3 | Pdzrn3 | 55983 | 9.1 (S) |
| Retinoic acid receptor, β | Rarb | 218772 | 4.7 (S) |
| Receptor tyrosine kinase-like orphan receptor 2 | Ror2 | 26564 | 3.0 (S) |
| Sal-like 3 (Drosophila) | Sall3 | 20689 | 3.2 (S) |
| Solute carrier family 6 (neurotransmitter transporter), member 14 | Slc6A14 | 56774 | 6.5 (S) |
| SPARC related modular calcium binding 1 | Smoc1 | 64075 | 4.8 (S) |
| Tachykinin receptor 3 | Tacr3 | 21338 | 13.6 (S) |
| Vav 3 oncogene | Vav3 | 57257 | 5.0 (S) |
| Zinc finger protein of the cerebellum 1 | Zic1 | 22771 | 3.7 (S) |
A sample of twenty genes from each of the Cochlea (C), Utricle (U), and Saccule (S) patterns of distinctive (signature) gene expression. Supplemental Table 4 lists all of the genes in these (and other) patterns that were upregulated by ≥1.5-fold.
The M and L categories (see Table 1) include substructures of the IE, such as the vestibular organ or cochlea. Therefore, these stages were separately analyzed by employing a two-factor analysis of variance test (ANOVA) to identify genes whose expression changed on the basis of developmental stage only, tissue type only, or on the basis of both developmental stage and tissue type. Only genes that met a P-value cutoff of ≤0.005 were considered for further analysis. An additional filter was then implemented using SAM to identify genes that changed by ≥1.5-fold with an estimated FDR of ≤0.5%.
For the middle (M) analysis only two patterns of tissue-specific differential expression were possible—genes high in the cochlea and those high in the vestibular organ (Table 2 and supplemental Figure 3C). Six patterns of gene expression were identified on the basis of developmental stage: genes high in E10.5 and E11 (“young” stages or Yng-M); those high in E11.5 and E12 (“old” stages or Old-M); genes up- or downregulated at E10.5 only (E10.5-Up and E10.5-Down, respectively); and genes up- or downregulated at E11 only (E11-Up and E11-Down, respectively).
In the late (L) analysis six possible tissue-specific patterns were identified; genes high in either one of the three dissected tissues or pairwise combinations of the three. Four patterns were identified on the basis of developmental stage; genes upregulated from E12.5 to E13.5 (young stages or Yng-L), those high from E14 to E15 (old stages or Old-L), and those up- or downregulated only at E14 (E14-Up and E14-Down, respectively). Both the M and the L analysis included comparisons to search for genes that changed in expression according to both tissue type and developmental stage. Genes within these groupings did not have a specific fold change associated with them. Instead, these were grouped according to their similarity of expression patterns using SOMs shown in supplemental Figure 4 (http://www.genetics.org/supplemental/). It should be noted that the above expression patterns are not always completely distinct from one another; some genes fall into more than one of the 28 patterns. This occurs because each of the three types of expression analyses (EML, M, and L) is a separate set of comparisons. Thus, there is no overlap of genes within one particular type of analysis. For example, genes within our EML analysis fall into unique, nonoverlapping patterns, but they may overlap with a pattern identified within the M analysis or within the L analysis. Thus, some genes recur when lists are compared between the three analyses.
The final step in this differential expression analysis was to compare all IE samples to their corresponding NIE control samples. That is, E9 IE was compared with E9 NIE; each IE sample from E9.5 to E10.5 was compared with NIE from the same stage; and each IE sample from E11 to E15 was compared with NIE from those stages. We identified genes that were either up- or downregulated in at least one IE sample relative to at least one NIE sample by twofold or more with an FDR of ≤0.5%. This resulted in the identification of 1410 genes that were upregulated and 226 genes that were downregulated in three or more (at least 10%) of all IE samples. Complete gene lists for the heat maps shown in supplemental Figure 3, for the SOMs in supplemental Figure 4, and for the IE vs. NIE comparisons are shown in supplemental Table 4.
GO classifications of differentially expressed genes:
To identify the functional categories represented by the various differentially expressed genes, we used GO annotations to classify these genes on the basis of molecular function (MF) ontology. Supplemental Table 5 (http://www.genetics.org/supplemental/) lists the significant MF classes and genes (along with their fold changes) that are unique to each type of expression pattern.
In the EML analysis (described above and in Tables 1 and 2), the highest number of differentially expressed genes was found in the EM expression pattern and, consequently, many significant MF terms are represented by these genes. This is not unexpected. During the early and middle developmental stages, when the vast majority of cells have not acquired their final differentiated states, it is likely that many different developmental routes will be elaborated with a consequently large number of gene expression changes. Some prevalent MF classes in this pattern were those involved in electron and proton transport, as well as helicase and DNA/histone binding activities. Genes high in the late stages included many coding for structural molecules unique to the IE [e.g., tectorins and collagens; TectA, TectB, Col18a1, and Col7a1 being upregulated in many IE samples relative to NIE ones (supplemental Table 4)], as well as those that bind certain growth factors and steroid hormones such as Nr1d2, Rarb, Rorc, and Vdr. Among the genes represented in the middle and late (ML) expression pattern were those encoding calcium and chloride ion channels and components of TGF-β signaling.
Within the M category, the vestibular organ, as expected, expressed many calcium ion binding proteins and various structural molecules. Numerous genes coding for zinc ion binding proteins were downregulated at E10.5 in both the cochlea and the vestibular organ (E10.5-Down) but these were dramatically upregulated after E10.5. These genes include Pcgf4, Rnf14, Rnf38, Zfp294, Pias1, and Pias2, among others. Interestingly, a similar burst of transcription from many zinc finger protein-coding genes has also been shown to occur as adult chicken auditory sensory epithelium undergoes regeneration following neomycin damage (Hawkins et al. 2007). It is tempting to speculate that this burst of avian zinc finger gene expression during hair cell regeneration may be a recapitulation of what normally occurs during development of the sensory epithelium.
In the L category, genes upregulated in the utricle included endopeptidase inhibitors, glutamate receptors, and potassium channels. Glutamate receptors are known to be expressed in the hair cells of the vestibular organ (Hendricson and Guth 2002), as are potassium channels (Eatock et al. 2002). Genes upregulated in the saccule included those encoding numerous zinc ion binding proteins. Interestingly, at E14 in all three tissues the transcription of many ribosomal protein genes was dramatically downregulated suggesting a possible reduction of protein synthesis throughout the IE at that stage. Certain transcriptional activators were also downregulated at this stage, indicating a possible slowing of overall transcriptional activity as well. Additionally, the transcription of many genes involved in energy production also appeared to be downregulated at this stage across all three organs. The E14-Up set of genes did not fall into significant and well-defined MF classes in this GO analysis (however, see below for pathway analysis on these and other genes) nor did genes within the Coch, Old-M, E11-Up, and C patterns.
Pathway and network analysis of differentially expressed genes:
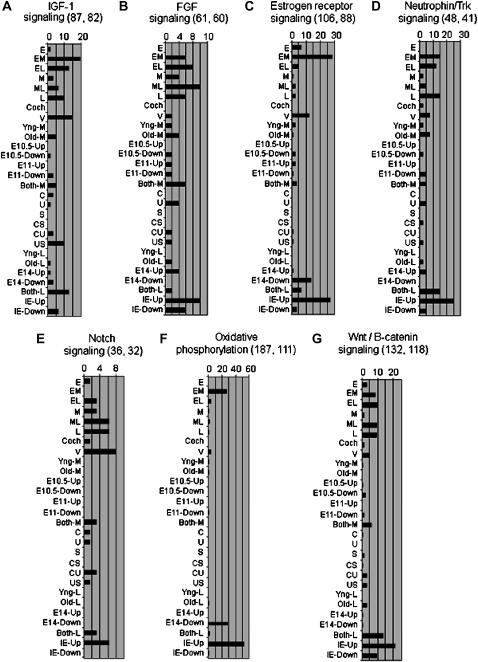
Gene ontologies provide a “broad brushstroke” view of functional annotations. However, we were interested in exploring which specific biological pathways and networks were represented within our data set. Therefore, we employed the Ingenuity Pathways Analysis (IPA) web-based series of tools (http://www.ingenuity.com) to analyze our data. IPA utilizes a database of manually curated relationships (direct and indirect) among human, mouse, and rat genes and gene products on the basis of original research publications from various scientific journals. This is known as the Ingenuity pathways knowledge base (IPKB). On the basis of this information, IPA creates molecular networks of direct physical, enzymatic, and transcriptional interactions defined by the genes uploaded by the user. We analyzed the lists of genes from each of the 28 expression patterns described above within the IPA suite of programs. Initially, we analyzed differentially expressed genes only with a fold change of at least 1.5-fold across tissues or time points. The constraints on this analysis were quite broad; we required only two genes within a given pathway to be present, but the output provides a different level of detail than GO annotations. We identified 53 Ingenuity signaling pathways that had two or more components differentially expressed in ≥1 of the 28 expression patterns. All of these various pathways are shown in supplemental Figure 5 (http://www.genetics.org/supplemental/). Supplemental Table 6 lists all of the genes parsed into all of the detected pathways and patterns of gene expression. Figure 4 shows seven examples of these pathways to illustrate what this type of analysis can reveal and how it differs from gene ontology outputs. Each of the 28 expression patterns is shown across the x-axes, and the y-axis for each pathway shows the percentage of genes from the pathway that are differentially expressed in each of the 28 patterns. The total number of genes in the pathway is shown at the top of each pathway, as is the total number present on the chip used in this study. Thus, it can be seen that 27% of the genes (classified by Ingenuity) within the estrogen receptor signaling pathway (Figure 4C) are differentially expressed during the early plus middle (EM) pattern of expression. This translates into a total of 24 genes (27% of 88 total pathway genes on the chip). Likewise, the vestibular (V) pattern of expression (upregulated in all vestibular samples relative to all cochlea samples) contains ∼11% (10 genes differentially expressed in total) of the total estrogen receptor signaling pathway. This pathway also appears to be downregulated at E14 (i.e., it is enriched in the E14 down pattern of expression), and it is enriched in the IE relative to the adjacent NIE tissues (IE-Up). Components of estrogen receptor signaling have been previously described in the developing and adult IE (Stenberg et al. 1999; Stenberg et al. 2002; also reviewed in Hultcrantz et al. 2006), but this analysis provides a much higher level of detail for this pathway.
Figure 4.—
Examples of pathways deemed statistically significant by IPA within at least one expression pattern resulting from at least one analysis (see Table 2). To be deemed significant, the pathway is represented by at least two genes differentially expressed by ≥2-fold (for patterns of EML analysis) or ≥1.5-fold (for patterns of M and L analyses) with a Fisher's right-tailed exact test P-value of ≤0.05. The horizontal axis shows the percentage of genes within the pathway that varied in expression within a particular pattern of gene expression. For each pathway (A–G), the total number of genes (listed by Ingenuity) in the pathway and the number of pathway genes actually on the gene chip are shown in parentheses next to the pathway name. For example, in Notch signaling (E) a total of 36 genes are listed by Ingenuity, of which 32 were on the gene chip. The M pattern of gene expression shows differential expression of 2 notch pathway genes (6.25% of 32 genes). The E pattern exhibits differential expression of only 1 notch pathway gene (3.1% of 32 genes) and is thus not deemed significantly enriched for this pathway. Refer to supplemental Figure 5 for a similar depiction of all 53 pathways identified in this study.
Figure 4, A, B, and D, shows components of the IGF-1, FGF, and Neurotrophin-Trk signaling pathways, respectively. All of these show quite broad representation through most of the 28 expression patterns. However, there are interesting and specific differences between patterns. For example, FGF signaling is enriched in both the IE-Up (overexpressed in IE vs. non-IE) and the IE-Down (overexpressed in NIE tissues). This apparently contradictory observation reflects the expression of different components of FGF signaling in the two tissue types. For example, Fgf9 is upregulated in the IE tissues whereas Fgf13 and Fgfr3 (among others) appear to be upregulated in the non-IE tissues.
The Notch signaling pathway (shown in Figure 4E) appears to be relatively enriched in the vestibular organs and in later stages of development. Specifically, genes such as Notch1, Notch2, and Notch4 appear to be higher in the vestibular (V) pattern of expression than in all of the cochlear samples. Notch1 is also one of the genes that is detectably upregulated in the later stages of development relative to the middle or early stages.
One of the most striking observations from the oxidative phosphorylation pathway shown in Figure 4F is that this pathway is over-represented by those genes that appear to be downregulated at E14 (E14-Down). This downregulation of housekeeping and metabolic genes at the E14 stage (already noted above) does not inversely correlate with an induction of specific, discernible IPA pathways in the E14-Up category. As noted above, the GO classifications for this set of genes are also not significantly enriched. Nevertheless, the E14-Up set of genes contains some interesting single genes that do show upregulation as proliferation apparently slows down. These genes include Intersectin1 (which is involved in endocytic membrane traffic) (Evergren et al. 2007), Fgfr1 (a key component of growth and differentiation in many systems, including the developing auditory sensory epithelium) (Pirvola et al. 2002), and Paxip1 (a member of the Pax gene family that is involved in maintaining genome stability) (Cho et al. 2003).
Wnt signaling is known to play a major role in IE development (Dabdoub et al. 2003; Stevens et al. 2003; Takebayashi et al. 2004; Kim et al. 2004; Ohyama et al. 2006). Numerous members of this (canonical) pathway were observed to have different expression patterns (Figure 4G) in the samples profiled. For instance, consistent with its role in cellular proliferation (Kim et al. 2004; Takebayashi et al. 2004), β-catenin (Ctnnb1) expression peaked in pattern EM (supplemental Table 4), at a time when the vast majority of cells in the IE are still undifferentiated and in a proliferative state. Many members of the frizzled and wnt family of genes also exhibited interesting patterns of expression. Specifically, within category L, Wnt7a appears to be upregulated by 3.1-fold in the cochlea relative to the saccule and the utricle, whereas Wnt4 is upregulated by 8.6-fold in both the cochlea and the saccule relative to the utricle. Within category M (before the saccule is morphologically distinct), Wnt4 expression changes on the basis of both tissue type and developmental stage, but it does so only in the cochlea. Hence, it appears that Wnt4 expression dramatically increases in the cochlea from E10.5 to E12 and then plateaus. Within the saccule Wnt4 expression increases as development progresses, but it does so linearly and does not show the plateau of expression observed in the developing cochlea. On average, Wnt4 expression was found to be 8.6-fold higher in all cochlear and saccular samples from E12.5 to E15 relative to the utricle samples. However, by the end of this time period (E15), Wnt4 expression was 6-fold higher in the cochlea and 17.4-fold higher in the saccule, compared to the utricle at the same stage. An additional wnt gene with an intriguing expression pattern is Wnt5a. Within category M, its expression is moderate and, on average, almost twice as high in the vestibular organ compared to the cochlea (supplemental Table 4). However, within category L Wnt5a expression increases sharply in the cochlea (but not in the utricle and the saccule) and peaks at E15 in this organ (supplemental Table 4 and centroids 5 and 64 in supplemental Figure 4B).
Genes from several immune-related pathways (such as natural killer cells, T- and B-cell signaling, and interleukin pathways) were also observed to significantly alter their expression in the profiled samples. These are listed in supplemental Table 6 and shown diagrammatically in supplemental Figure 5. In all three analyses, most of these pathways were found to be statistically significant in expression patterns on the basis of developmental stage, suggesting common roles in various organs of the IE. It is known that certain immune cells are present in the mature IE and are recruited to sites of hair cell damage in both the cochlea and vestibular organs of mammals and birds (Fredelius and Rask-Andersen 1990; Warchol 1997, 1999; Bhave et al. 1998; Warchol and Kaplan 1999). Studies also indicate that immune cells can assist in wound healing (Brown et al. 1993; Hubner et al. 1996; Warchol et al. 2001) by phagocytosing debris from dead cells and secreting cytokines and other molecules that promote recovery. The observation that immune pathways are active even during early development of the mouse IE indicates that these may function either to promote cellular proliferation or remove debris (or both) produced as a result of the normal programmed cell death (PCD) in the developmental process. The latter has been well documented beginning from the otic cup stage to shortly after birth (Fekete et al. 1997; Cecconi et al. 2004; Leon et al. 2004). Various caspases and Bcl genes, known PCD participants, were observed to significantly alter in their expression. For instance, Bcl2l1 and caspases-3, -6, and -8 followed the expression pattern EM, whereas Casp7 and Bcl2 fell into patterns ML and L, respectively. Within category L, the pro-apoptotic gene Bid was upregulated in the cochlea and saccule relative to the utricle, while Casp9 was upregulated in the utricle and saccule relative to the cochlea (supplemental Table 4). Bcl2, on the other hand, increased sharply in the cochlea from E12.5 and peaked at E15 in this organ, but not in the saccule and utricle (supplemental Table 4 and centroid 65 in supplemental Figure 4B).
Evaluating relationships among genes through network building:
The analysis illustrated in Figure 4 and in supplemental Figure 5 identifies a series of seed networks in which differentially expressed genes resided. However, these networks include only genes that are detectably differentially expressed. They do not take into account genes that are not differentially expressed, but are in fact present across various time points and tissues and are part of the network. We were interested in determining whether these “missing” genes were actually present in a given tissue or stage, but were not scored as being differentially expressed across the sample set. Therefore, we queried all genes within our seed networks for whether they were scored as “present” in IE samples comprising a particular expression pattern (e.g., early genes). We took all those that were present, combined them with the differentially expressed genes and re-uploaded this set into IPA to determine if they further populated the initial pathway or others. In this way we derived the types of network outputs shown in Figure 5. This second form of analysis consists of a series of interaction diagrams in which networks of direct and indirect gene interactions can be postulated from the IPKB. In many cases these are large in scale and should provide many new leads into the exact framework of interactions that occur during the complex process of IE development and morphogenesis.
Figure 5.—
Examples of networks generated by IPA using genes whose expression was upregulated in (A) L (late category), (B) C (late cochlea), and (C) S (late saccule). Each gene list was uploaded in Ingenuity to identify interactions among the genes within the list and also with other genes not in the list. Upregulated genes are shown in different shades of red (fold change increases from light to bright red), and those in green are genes that were not differentially expressed but were present in the appropriate category/subcategory (L, C, or S). Solid blue lines denote direct interactions and dashed blue lines denote indirect interactions (refer to key). Also indicated are genes that are parts of several biological signaling pathways with a known role in the IE.
The network in Figure 5A was generated by merging two of the high-scoring networks built by IPA using genes upregulated in category L (shown as varying shades of red, proportional to their level of expression) by at least 1.5-fold relative to categories E and M. Genes shown in green are those that are known to interact with upregulated genes, but which themselves were not upregulated in this category. However, they were present within this category. Thus, one can see that seven components of Wnt signaling are enriched in these late stages, as are interacting components of Notch signaling, components of the Ap1 pathway, and various components that interact with the hair cell differentiation marker Atoh1. A total of 68 genes are thus drawn into this network.
Figure 5B shows an example of one of several high-scoring networks built around genes upregulated in the cochlea (again shown in shades of red) by at least 1.5-fold within category L relative to both the utricle and the saccule, while those in green are known to interact with the former and were found to be present in the late cochlear samples. Genes from three pathways (TGF-β, Vegf, and NF-κB) known for their roles in IE development are indicated on this figure. In addition to these, it is interesting to note that additional upregulated genes, such as Irx5 and Clu, are also found within this network (black arrows). It appears likely that these genes (which have not previously been investigated in the IE and were validated by in situs, see below) function through one or more of the three known pathways within the cochlea at later stages in IE development.
The network in Figure 5C was built similarly to those in 5A and 5B but using genes upregulated in the saccule by at least 1.5-fold (shades of red) within category L relative to the cochlea and the utricle. In this case, in addition to genes from well known IE pathways (e.g., Wnt and the cell cycle), there are also genes (e.g., Clock and Per2) from the circadian rhythm pathway. There is evidence that appears to link the vestibular system to homeostatic and circadian regulation (Fuller et al. 2002), but this pathway has not been characterized in the IE. It is interesting to note that this pathway has also been observed to be differentially expressed in regenerating avian sensory epithelia (Hawkins et al. 2007).
RNA in situ hybridization confirms differential expression:
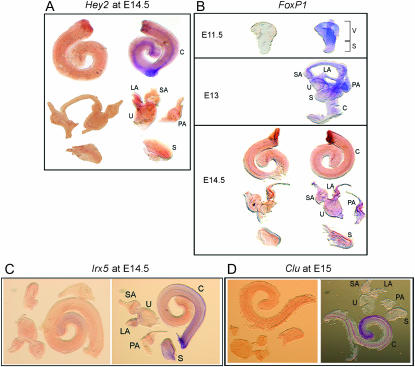
To validate our array data and also to identify spatial patterns of expression, we performed whole mount RNA in situ hybridizations on microdissected IE structures. Genes for this analysis were selected from the array data on the basis of their apparent clear-cut, tissue-specific patterns of gene expression and the fact that they had never previously been studied in the IE. On the basis of these criteria we selected hairy and enhancer of split related with YRPW motif 2 (Hey2, a gene that appeared from our array data to be cochlea-specific at E14.5), iroquois homeobox protein 5 (Irx5, another gene that appeared to be cochlea-specific), forkhead box protein1 (FoxP1, which showed a vestibular pattern of expression), and Clusterin (Clu, which showed a cochlear-specific pattern of expression).
Hey2 expression was observed to be localized in a strip of cells running down the middle of the cochlear duct at E14.5 (Figure 6A) corresponding to the location of the developing sensory epithelium, but was not detectable in vestibular organs. This tissue specificity is entirely in agreement with the array data. Given that two other hairy and enhancer of split genes, Hes1 and Hes5, are known to be negative regulators of hair cell differentiation and are localized in supporting cells (Zheng et al. 2000; Zine et al. 2001), it is possible that Hey2 functions similarly in the IE. In this regard, it is intriguing that we detected a 3-fold higher level of Hey2 expression in the cochlea compared to the utricle and saccule, whereas expression of Hes5 was 8.7-fold higher in the utricle compared to the cochlea and the saccule. This latter observation is consistent with previous RNA in situ data for Hes5 (Shailam et al. 1999), which demonstrated that it is expressed only in the cristae of the three semicircular canals during these stages of development. Given that in the current study the utricle was profiled together with the three ampullae (which contain the cristae), the source of high expression signal for Hes5 in this mixture would be entirely from the latter structures. In our array data Hes5 was not detectably expressed in the cochlea until E15 and was absent in all samples of the saccule, again consistent with previous reports (Shailam et al. 1999; Zine et al. 2001).
Figure 6.—
Confirmatory whole mount RNA in situ hybridizations using antisense (right) and control sense (left) riboprobes for genes that show distinct patterns of gene expression from gene chip analysis. (A) Hey2 in the cochlea at E14.5 is localized in a stripe of cells that runs through the middle of the organ and corresponds to the region of the developing sensory epithelium. Its expression is not detectable in the components of the vestibular organ. See supplemental Figures 7 and 8 (http://www.genetics.org/supplemental/) for a higher resolution view. (B) FoxP1 at E11.5, E13, and E14.5. FoxP1 transcripts are significantly overexpressed in the vestibular region at E11.5 and at E13 this intensifies within the utricle and the three ampullae. This gene is also expressed within the semicircular canals, but at a reduced expression level. At E14.5 the message is still detected in the utricle and all three ampullae, although at a lower level compared to that at E13. However, expression of this gene is not detectable in the cochlea and in the saccule. Supplemental Figures 9–11 (http://www.genetics.org/supplemental/) provide a higher resolution view. (C) Irx5 at E15 appears to be localized in the cochlea, with a diffuse expression detected in the saccule as well. Within the cochlea it is expressed in a gradient that decreases from the base to the apex, with a more intense signal being detected in a stripe of cells along the very outer edge of this organ's curve. Note that the strong staining at the basal tip is nonspecific. See supplemental Figures 12 and 13 for a higher resolution view. (D) Clusterin at E15 is detected only in the cochlea and is expressed in two stripes that begin at the base of the cochlea and fuse at its apex. It appears to be expressed in a gradient opposite to that of Irx5 in that it is upregulated at the apex rather than at the base. For higher resolution images refer to supplemental Figures 14 and 15.
Transcripts for Irx5 are localized in the cochlea (Figure 6B), as suggested by the array data, and are present in a gradient that decreases from the base to the apex. The saccule also appears to express Irx5, although in a diffuse pattern. Iroquois genes frequently have overlapping domains of expression during development, and appear to play redundant roles (Bosse et al. 1997; Bruneau et al. 2001; Houweling et al. 2001; Lebel et al. 2003). Interestingly, our array data indicate that three additional iroquois genes (Irx1, Irx3, and Irx6) are also upregulated in the cochlea by 2.2-, 9.5-, and 1.8-fold, respectively, relative to the utricle and saccule. Irx5 is involved in cone bipolar cell differentiation in the mouse retina (Cheng et al. 2005) and also in maintaining the ventricular repolarization gradient in the heart by repressing the expression of Kcnd2, a potassium channel gene (Costantini et al. 2005). On the basis of these observations in the heart, it is worth noting that Kcnd2 expression is upregulated in the utricle by 22.9-fold relative to the cochlea and saccule (over the same time period as Irx5) suggesting that Irx5 might play a similar role in the IE.
FoxP1 expression was found to be vestibular specific, as reported by the array data, and was localized to the utricle and the ampullae of the three semicircular canals (Figure 6C). This gene is known for its role in cardiac development (Wang et al. 2004) and also in the differentiation of monocytes where it binds the promoter of the gene coding for the macrophage colony stimulating factor receptor (Csf1r) and represses its expression (Shi et al. 2004). The expression of FoxP1 itself in monocytes is initiated as a result of clustering of a membrane-bound integrin Itgam. Furthermore, lack of FoxP1 also results in defective B-cell development and a reduction in the levels of Rag1 and Rag2 proteins that are involved in V(D)J recombination in these cells because FoxP1 directly controls their expression (Hu et al. 2006). There is a resident population of immune cells in the IE (Warchol 1997; Bhave et al. 1998), and it is possible that FoxP1 is involved in their differentiation. However, the fact that its expression is localized in the utricle and the three ampullae suggests a more specific role for this gene in these particular sensory structures of the IE. Another forkhead gene detected in our array data, FoxG1, was found to be overexpressed in the late cochlea by 6.3-fold relative to the utricle and saccule, even though it was still present in the latter two organs. This is consistent with previous studies in which mice lacking this gene were found to have several IE abnormalities including a shortened cochlea with several rows of hair and support cells, defective innervation in both the cochlea and the vestibule, and absence of lateral crista (Pauley et al. 2006).
Clusterin (Clu) also known as ApoJ, has been shown to have numerous roles, which include influencing the deposition of β-amyloid in the brain (Demattos et al. 2004), protecting heart tissue from postinflammatory tissue destruction (Mclaughlin et al. 2000), acting as a tumor suppressor by inhibiting the NF-κB pathway (Santilli et al. 2003), the activity of which is required for tumor invasion, and being a heat-shock protein with chaperone activity (Wilson and Easterbrook-Smith 2000). RNA in situ (Figure 6D) revealed that it is expressed only in the cochlea in two stripes of cells running down the cochlear duct that fuse at the apex. These appear to correspond to the sensory epithelia and the stria vascularis. It is unclear what specific role this gene might play in the IE, but in the context of this study it is interesting to note that it is cochlear specific in agreement with the array data.
In addition to the RNA in situs for four genes presented here, supplemental materials also include data for four additional genes, all of which support the expression patterns observed with microarrays: Kcnd2 [supplemental Figure 16 (http://www.genetics.org/supplemental/)], Ttyh1 (supplemental Figure 17), Slc2a3 (supplemental Figures 18–20), and Zbtb16 (supplemental Figure 21). A semiquantitative RT–PCR method was also employed as another independent method of validating microarray observations on eight additional genes: Irf6, Rxrg, Slc2a3, Lin28, Ttyh1, Zfp503, Fst, and Pthlh (note that for two of these genes, Slc2a3 and Ttyh1, RNA in situs were also carried out). Supplemental Figure 22 and supplemental Table 9 include this PCR data. The microarray and PCR data show excellent agreement in the observed patterns of gene expression. These independent methods of assessing gene expression not only serve to validate our array data, but also illustrate how this data set can readily provide numerous new, interesting, spatially and temporally regulated genes to further investigate during IE development.
Candidate genes for human deafness loci:
Currently, >110 human genomic intervals have been identified that harbor nonsyndromic deafness loci (the Hereditary Hearing Loss home page, http://webh01.ua.ac.be/hhh/). In many cases the pedigrees from which these locations were identified are quite small, the genomic intervals are large and contain many candidate genes. Consequently, only 40 of these loci have been characterized at the mutational level. A similar situation exists in the mouse where, despite the ability to narrow intervals by repeated crosses, many deafness/balance loci remain uncloned (http://www.jax.org/hmr/map.html). Therefore, we sought to determine how many of our differentially expressed genes fell within as yet uncloned deafness genetic intervals and might thus be considered candidate genes. Table 5 lists a sampling of 10 human genetic intervals that contain mouse orthologs differentially expressed within the IE samples as well as those that were upregulated specifically in more than one-third of all IE samples relative to NIE ones. Supplemental Table 8 (http://www.genetics.org/supplemental/) lists all 54 human genetic intervals that contain IE-expressed orthologs (in addition to the differentially expressed ones) found in this study. Supplemental Table 8 also lists the estimated size of each published interval and the number of genes within each interval (from the UCSC genome browser, http://genome.ucsc.edu/). The estimated interval sizes range from just under 1 Mb to >31 Mb, and the total number of genes in the target intervals ranges from 5 to >380. The total number of genes found to be differentially expressed per interval ranges from 1 to 82 and roughly correlates with the total number of genes per interval. There are nine intervals that have three or fewer differentially expressed candidate genes within them. Hopefully, these data will prove useful to many groups of investigators in resolving exactly which mutations cause these various inherited forms of deafness.
TABLE 5.
Examples of 10 nonsyndromic human deafness intervals for which no causative gene has been identified to date, and candidate genes in these intervals identified in the current study
| Genes differentially expressed by ≥1.5-fold | Interval (size in Mb) | Markers | Total no. of genes in interval |
|---|---|---|---|
| ACTR10, C14ORF100 | DFNA23 (8.03) | D14S980–D14S1046 | 79 |
| DACT1, HIF1A, HSPA2 | |||
| MNAT1, MTHFD1, OTX2a | |||
| PPM1A, PRKCH, PSMA3 | |||
| RHOJ, RTN1, SIX1a | |||
| TIMM9, TMEM30B | |||
| AP3S2, BLM, CIB1a | DFNA30 (7.11) | D15S151–D15S130 | 74 |
| FURIN, IQGAP1 | |||
| LOC400451, MRPL46 | |||
| MRPS11, NTRK3 | |||
| PEX11A, POLG, PRC1 | |||
| RLBP1, VPS33B | |||
| CHFR, DDX51, EP400 | DFNA41 (4.05) | D12S1609-tel | 48 |
| GOLGA3, POLE, PXMP2, RANa | |||
| ANKHD1, APBB3, BRD8 | DFNA42 (11.99) | D5S2056–D5S638 | 172 |
| C5ORF5, CDC23, CDC25C | |||
| CTNNA1a, CXCL14, CXXC5 | |||
| EGR1, ETF1, GNPDA1 | |||
| H2AFY, HARS, HARSL | |||
| HSPA9B, IK, KIAA0141 | |||
| KIF20A, LOC340061 | |||
| MATR3, NDFIP1, NDUFA2 | |||
| NME5, ORF1-FL49, PCDHB13 | |||
| PCDHB17, PCDHB9, PFDN1 | |||
| POU4F3, PPP2R2B, PURA | |||
| RNF14, SIL1, SLC35A4 | |||
| SMAD5, SRA1, TAF7 | |||
| TCERG1, TGFBI, UBE2D2 | |||
| C14ORF21, DHRS1 | DFNA53 (6.97) | D14S581–D14S1021 | 60 |
| DHRS4, FOXG1Ba | |||
| GMPR2, IPO4, ISGF3G | |||
| NEDD8, NFATC4, PRKCM | |||
| PSME2, RABGGTA, SCFD1 | |||
| STXBP6, TINF2, WDR23 | |||
| ARPC1B, ARS2, ASNS | DFNB14 (11.93) | D7S527–D7S3074 | 172 |
| ATP5J2, BCAP29, BRI3 | |||
| CBLL1, COPS6a, CPSF4 | |||
| CUTL1, CYP3A5 | |||
| DKFZP434B0335, DKFZP434K1815 | |||
| DLX5, EMID2, EPHB4 | |||
| GNB2, HBP1, HRBL | |||
| LAMB1, LRCH4, MCM7 | |||
| MLL5, ORC5L, PBEF1 | |||
| PCOLCE, PDAP1, PIK3CG | |||
| PILRB, POLR2Ja, POP7 | |||
| PSMC2, PTCD1, SHFM1 | |||
| SLC12A9, SLC25A13a | |||
| SLC26A4a, SRPK2, SVH | |||
| SYPL, TAC1, TAF6, ZNF3 | |||
| ZNF38, ZNF394, ZNHIT1 | |||
| BCAP29, CBLL1, DNAJB9 | DFNB17 (3.99) | D7S2453–D7S525 | 24 |
| HBP1, IPLA2(GAMMA) | |||
| LAMB1, NRCAM, PBEF1 | |||
| PIK3CG, SLC26A4a, SYPL | |||
| ABCD4, ACYP1, ALDH6A1 | DFNB35 (7.85) | D14S588–D14S59 | 110 |
| C14ORF112, C14ORF133 | |||
| C14ORF169, CHX10, EIF2B2 | |||
| ENTPD5, ESRRB, FOS, GSTZ1 | |||
| JDP2, MED6, NEK9, NPC2 | |||
| NUMB, PCNX, PGF, POMT2 | |||
| PSEN1, RBM25, SMOC1a | |||
| SYNJ2BP, TGFB3 | |||
| AUTS2, BAZ1B, CACNA2D1 | DFNB39 (17.93) | D7S3046–D7S644 | 114 |
| CALN1, CLDN3a, CLDN4a | |||
| ELN, FZD9, GNAI1, GTF2I | |||
| GTF2IRD1, GTF2IRD2, HGF | |||
| HIP1a, LIMK1, LOC54103 | |||
| MDH2, PCLO, PHTF2, POM121 | |||
| PTPN12, SEMA3A, SEMA3E | |||
| STX1A, WBSCR1, YWHAG | |||
| ADAMTS7, ARID3B, ARIH1a | DFNB48 (11.66) | D15S216–D15S1041 | 155 |
| CIB2, COX5Aa, CRABP1 | |||
| CSPG4, CTSH, ETFA, FAH | |||
| HEXA, HMG20A, IDH3A | |||
| ISL2, ISLR, MAN2C1 | |||
| MORF4L1, MTHFS, NEO1 | |||
| PKM2, PTPN9, RASGRF1 | |||
| SCAMP2, SCAMP5, SDFR1 | |||
| SIN3A, STARD5, TLE3 |
The human nonsyndromic deafness intervals shown here contain mouse orthologs found in this study to be differentially expressed (listed in the first column). Refer to supplemental Table 8 for a listing of all 54 intervals.
Genes upregulated in at least 30% of all IE samples relative to at least one NIE sample.
DISCUSSION
In this article we describe the most detailed gene expression profiles and pathway information to date over the course of mouse IE organogenesis. Particular care was taken to derive high quality biological samples and to derive statistically robust expression profiles from a strain of mice (CBA/J) that does not exhibit a significant age-related hearing loss (Hunter and Willott 1987). We collected samples at half-day intervals starting at one of the earliest stages of IE development (the E9 otic cup) and separately dissected substructures up to E15, when the specialized sensory epithelia in all six sensory organs have already begun to differentiate. By profiling in duplicate 29 IE and three NIE tissues (i.e., a total of 64 individual arrays) we have obtained a first glimpse into the “tool box” of gene expression changes that specify the development of the complex cell types and structures of the IE. Our data are freely available through the National Center for Biotechnology Information's GEO database, and we have presented extensive analyses (including RNA in situ data) that validate their quality.
In addition to describing the derivation and quality of the data, we have attempted to analyze this large data set in a manner that makes it useful and accessible to other investigators. One way that we have approached this is by mapping IE genes into as yet uncloned deafness intervals, as described above, to assist many ongoing positional cloning projects. However, our major efforts have focused upon a set of pairwise comparisons of all stages and tissues to identify temporal and/or tissue-specific patterns of gene expression. In general, we did not observe many dramatic and absolute changes in gene expression (the only clear-cut exception being seven genes in the E15 cochlea). Instead, we observed a more nuanced pattern of changes occurring across multiple time points or tissue types. In many cases particular genes exhibited large fold changes in gene expression, but the expression trends were spread over several time points or tissue types. We conducted four types of comparative analysis (EML, M, L, and IE vs. NIE) to identify 28 patterns of gene expression. By pathway analysis we further identified 53 signaling cascades that were enriched within these expression patterns. Many of these 53 pathways were shared between the 28 different expression patterns. This does not necessarily reflect the repeated use of the same pathway components in each of the gene expression patterns. Instead, individual genes within these pathways have unique patterns of expression, indicating that specific components of these pathways are utilized at particular times and/or in certain tissues. For example, multiple components of Wnt signaling [which is known to play a role during IE development (Dabdoub et al. 2003; Stevens et al. 2003; Kim et al. 2004; Takebayashi et al. 2004; Ohyama et al. 2006)] occur in the majority of the 28 expression patterns. However, different components of this pathway are expressed at particular stages of development. For example, four genes from the Wnt pathway are upregulated only in early stages and eleven are upregulated only in late stages of IE development (supplemental Table 6). An additional example of this differential use of specific components is provided by the G1/S-phase checkpoint pathway of the cell cycle. At first glance, the cyclin-dependent kinase inhibitor genes Cdkn1B (p27Kip1), Cdkn2D (p19Ink4d), and Cdkn2B (p15Ink4b) are all upregulated in the late pattern of gene expression. This is to be expected, especially given that the first two are known to be expressed during this late time period and maintain the postmitotic state of differentiated cochlear hair cells (Chen and Segil 1999; Chen et al. 2003; Lee et al. 2006). However, according to our data set the actual expression patterns of these genes within the three tissues during late development (i.e., in the cochlea, saccule, and utricle) are not identical. Expression of Cdkn1B dramatically increases in the cochlea (but not in the utricle or saccule) at E12.5 and peaks at E15. This expression pattern is consistent with previous immunostaining studies of the cochlea during this time period (Lee et al. 2006). On the other hand, Cdkn2D expression increases almost linearly in the saccule from E12.5 to E15. By E15, expression is 4.6 times higher in the saccule and 2.6 times higher in the utricle relative to the E15 cochlea. Cdkn2B expression also appears to vary by tissue during this time frame; it is highly expressed in the saccule compared to the cochlea and utricle. These specific observations are possibly due to differences in the control of cell cycle exit and the subsequent maintenance of the quiescent state of hair and supporting cells in different organs of the IE. The general theme underlying all of these differences in gene expression is that different components of particular pathways are employed in different places and at different times during IE development, and that these subtleties can be identified within our data.
Many of the pathways we have identified involve genes that are known to play key roles in IE development (e.g., Wnt, Notch, and Fgfs). Our analysis not only has added many other genes that act within these pathways into our understanding of the process, but also has revealed several unexpected pathways and patterns. Examples of these are the circadian rhythm and estrogen receptor signaling pathways (among others). It is unclear what role these pathways might play in ear development. Expression of CLOCK family genes is clearly not limited to regions of the nervous system that govern circadian rhythms. Cells in most peripheral tissues also contain endogenous oscillators on the basis of this same genetic network (Oishi et al. 2003; Yamamoto et al. 2004; Tsinkalovsky et al. 2006). However, differential expression of this network is a novel observation in IE development. Additionally, while estrogen receptor expression has been previously observed in the adult mouse IE (Stenberg et al. 1999; Stenberg et al. 2002), our observation that multiple components of this signaling pathway are differentially expressed in the developing IE is novel. It is interesting to note that in a separate study of gene expression in avian IE sensory epithelial regeneration we also observed changes in estrogen receptor signaling, including estrogen receptor α. It is unclear what role this pathway is playing in either of these systems, but it is known from other systems that activation of the pathway does not necessarily require estrogen as a ligand. Ligand-independent activation of ER can be achieved by ER phosphorylation mediated by various other signaling pathways and signaling molecules (Sommer and Fuqua 2001).
One surprising pattern of gene expression we observed was the coordinate downregulation of numerous genes involved in protein synthesis, i.e., constituents of ribosomes (∼50 genes, see supplemental Table 5) and oxidative phosphorylation (∼30 genes, see supplemental Tables 5 and 6) in all structures profiled at E14. This suggests that some form of overall regulatory control might underlie this coordinate change in transcripts that are so integral to cellular growth and energy metabolism. These changes happen during a period at which many IE differentiation events are occurring, including the differentiation of sensory and nonsensory cells in the cochlea and vestibular organs. It is possible that the apparent downregulation of so many metabolic genes is in fact a reflection of the decreased proliferation and lowered energy requirements of these differentiating cells during this time period. The immediate increase in these transcripts beyond E14 may be the result of an increase in the energy demands of specialized structures as they mature and become innervated.
Clearly, our data add a large number of interesting genes and pathways to the list of those involved in IE development. We have also identified gene expression signatures for particular IE structures and/or stages of development. These clear-cut patterns of gene expression provide diagnostic gene expression “bar codes” for IE development. They represent gene expression changes occurring in the sampled structure at that particular point in development and are thus a reflection of all the regulatory interactions occurring in that time and place. As IE biologists proceed further with genomic approaches for the analysis of smaller structures and specific cell types within the IE, it is to be hoped that these larger signatures can eventually be deconstructed into a series of underlying gene expression patterns and specific interactions that will help to solve the genetic “wiring diagram” of this important organ.
Acknowledgments
The authors are grateful to Anne Bowcock for her critical reading of this manuscript. This work was supported by grant no. RO1DC5632 from the National Institute on Deafness and Other Communication Disorders (to M.L.).
Sequence data from this article have been deposited with the National Center for Biotechnology Information's Gene Expression Omnibus under series accession no. GSE7536.
References
- Anagnostopoulos, A. V., 2002. A compendium of mouse knockouts with inner ear defects. Trends Genet. 18: 499. [DOI] [PubMed] [Google Scholar]
- Anniko, M., 1983. Cytodifferentiation of cochlear hair cells. Am. J. Otolaryngol. 4: 375–388. [DOI] [PubMed] [Google Scholar]
- Avraham, K. B, 2003. Mouse models for deafness: lessons for the human inner ear and hearing loss. Ear Hear. 24: 332–341. [DOI] [PubMed] [Google Scholar]
- Beisel, K. W., T. Shiraki, K. A. Morris, C. Pompeia, B. Kachar et al., 2004. Identification of unique transcripts from a mouse full-length, subtracted inner ear cDNA library. Genomics 83: 1012–1023. [DOI] [PubMed] [Google Scholar]
- Bhave, S. A., E. C. Oesterle and M. D. Coltrera, 1998. Macrophage and microglia-like cells in the avian inner ear. J. Comp. Neurol. 398: 241–256. [DOI] [PubMed] [Google Scholar]
- Bosse, A., A. Zulch, M-B. Becker, M. Torres, J. L. Gomez-Skarmeta et al., 1997. Identification of the vertebrate Iroquois homeobox gene family with overlapping expression during early development of the nervous system. Mech. Dev. 69: 169–181. [DOI] [PubMed] [Google Scholar]
- Brown, L. F., D. Dubi, L. Lavigne, B. Logan, H. F. Dvorak et al., 1993. Macrophages and fibroblasts express embryonic fibronectins during wound healing. Am. J. Pathol. 142: 793–801. [PMC free article] [PubMed] [Google Scholar]
- Bruneau, B. G., Z. Z. Bao, D. Fatkin, J. Xavier-Neto, D. Georgakopoulos et al., 2001. Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol. Cell. Biol. 21: 1730–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarero, G., C. Avendano, C. Fernandez-Moreno, A. Villar, J. Contreras et al., 2001. Delayed inner ear maturation and neuronal loss in postnatal Igf-1-deficient mice. J. Neurosci. 19: 7630–7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantos, R., L. K. Cole, D. Acampora, A. Simeone and D. K. Wu, 2000. Patterning of the mammalian cochlea. Proc. Natl. Acad. Sci. USA 97: 11707–11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi, F., K. A. Roth, O. Dolgov, E. Munarriz, K. Anokhin et al., 2004. Apaf1-dependent programmed cell death is required for inner ear morphogenesis and growth. Development 9: 2125–2135. [DOI] [PubMed] [Google Scholar]
- Chen, P., and N. Segil, 1999. p27Kip1 links cell proliferation to morphogenesis in the developing organ of Corti. Development 126: 1581–1590. [DOI] [PubMed] [Google Scholar]
- Chen, P., F. Zindy, C. Abdala, F. Liu, X. Li et al., 2003. Progressive hearing loss in mice lacking the cyclin-dependent kinase inhibitor Ink4d. Nat. Cell Biol. 5: 422–426. [DOI] [PubMed] [Google Scholar]
- Chen, Z-Y., and D. P. Corey, 2002. An inner ear gene expression database. J. Assoc. Res. Otolaryngol. 3: 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C. W., R. L. Chow, M. Lebel, R. Sakuma, H. O. Cheung et al., 2005. The Iroquois homeobox gene, Irx5, is required for retinal cone bipolar cell development. Dev. Biol. 287: 48–60. [DOI] [PubMed] [Google Scholar]
- Cho, E. A., M. J. Prindle and G. R. Dressler, 2003. BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol. Cell. Biol. 23: 1666–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini, D. L., E. P. Arruda, P. Agarwal, K. H. Kim, Y. Zhu et al., 2005. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell 21: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub, A., M. J. Donohue, A. Brennan, V. Wolf, M. Montcouquiol et al., 2003. Wnt signaling mediates reorientation of outer hair cell stereociliary bundles in the mammalian cochlea. Development 130: 2375–2384. [DOI] [PubMed] [Google Scholar]
- Dechesne, C. J., and M. Thomasset, 1988. Calbindin (CaBP 28 kDa) appearance and distribution during development of the mouse inner ear. Brain Res. 468: 233–242. [DOI] [PubMed] [Google Scholar]
- Demattos, R. B., J. R. Cirrito, M. Parsadanian, P. C. May, M. A. O'Dell et al., 2004. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron 41: 193–202. [DOI] [PubMed] [Google Scholar]
- Eatock, R. A., K. M. Hurley and M. A. Vollrath, 2002. Mechanoelectrical and voltage-gated ion channels in mammalian vestibular hair cells. Audiol. Neurootol. 7: 31–35. [DOI] [PubMed] [Google Scholar]
- Evans, A. L., and U. Muller, 2000. Stereocilia defects in the sensory hair cells of the inner ear in mice deficient in integrin alpha8beta1. Nat. Genet. 4: 424–428. [DOI] [PubMed] [Google Scholar]
- Evergren, E., H. Gad, K. Walther, A. Sundborger, N. Tomilin et al., 2007. Intersectin is a negative regulator of dynamin recruitment to the synaptic endocytic zone in the central synapse. J. Neurosci. 27: 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete, D. M., S. A. Homburger, M. T. Waring, A. E. Riedl and L. F. Garcia, 1997. Involvement of programmed cell death in morphogenesis of the vertebrate inner ear. Development 124: 2451–2461. [DOI] [PubMed] [Google Scholar]
- Fredelius, L., and H. Rask-Andersen, 1990. The role of macrophages in the disposal of degeneration products within the organ of corti after acoustic overstimulation. Acta Otolaryngol. 109: 76–82. [DOI] [PubMed] [Google Scholar]
- Fritzsch, B., K. F. Barald and N. I. Lomax, 1998. Embryology of the vertebrate ear, pp. 80–145 in Development of the Auditory System, edited by E. W. Rubel, A. N. Popper and R. R. Far. Springer, New York.
- Fuller, P. M., T. A. Jones, S. M. Jones and C. A. Fuller, 2002. Neurovestibular modulation of circadian and homeostatic regulation: Vestibulohypothalamic connection? Proc. Natl. Acad. Sci. USA 99: 15723–15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, B. C., J. H. Jonathan and R. M. Grainger, 1996. Inductive processes leading to inner ear formation during Xenopus development. Dev. Biol. 175: 95–107. [DOI] [PubMed] [Google Scholar]
- Golub, T. R., D. K. Slonim, P. Tamayo, C. Huard, M. Gaasenbeek et al., 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286: 531–537. [DOI] [PubMed] [Google Scholar]
- Hawkins, R. D., S. Bashiardes, C. A. Helms, L. Hu, N. L. Saccone et al., 2003. Gene expression differences in quiescent versus regenerating hair cells of avian sensory epithelia: implications for human hearing and balance disorders. Hum. Mol. Genet. 12: 1261–1272. [DOI] [PubMed] [Google Scholar]
- Hawkins, R. D., C. A. Helms, J. B. Winston, M. E. Warchol and M. Lovett, 2006. Applying genomics to the avian inner ear: development of subtractive cDNA resources for exploring sensory function and hair cell regeneration. Genomics 87: 801–808. [DOI] [PubMed] [Google Scholar]
- Hawkins, R. D., S. Bashiardes, K.E. Powder, V. Bhonagiri, D.M. Alvarado et al., 2007. Large scale gene expression profiles of regenerating inner ear sensory epithelia. PLoS ONE 2: e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricson, A. W., and P. S. Guth, 2002. Transmitter release from Rana pipiens vestibular hair cells via mGluRs: a role for intracellular Ca(++) release. Hear. Res. 172: 99–109. [DOI] [PubMed] [Google Scholar]
- Houweling, A., R. Dildrop, T. Peters, J. Mummenhoff, A. F. M. Moorman et al., 2001. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech. Dev. 107: 169–174. [DOI] [PubMed] [Google Scholar]
- Hu, H., B. Wang, M. Borde, J. Nardone, S. Maika et al., 2006. Foxp1 is an essential transcriptional regulator of B cell development. Nat. Immunol. 7: 819–826. [DOI] [PubMed] [Google Scholar]
- Hubner, G., M. Brauchle, H. Smola, M. Madlener, R. Fassler et al., 1996. Differential regulation of pro-inflammatory cytokines during wound healing in normal and glucocorticoid-treated mice. Cytokine 8: 548–556. [DOI] [PubMed] [Google Scholar]
- Hultcrantz, M., R. Simonoska and A. E. Stenberg, 2006. Estrogen and hearing: a summary of recent investigations. Acta Otolaryngol. 126: 10–14. [DOI] [PubMed] [Google Scholar]
- Hunter, K. P., and J. F. Willott, 1987. Aging and the auditory brainstem response in mice with severe or minimal presbycusis. Hear. Res. 30: 207–218. [DOI] [PubMed] [Google Scholar]
- Kelley, M. W., D. K. Wu, A. N. Popper and R. R. Fay, 2005. Development of the Inner Ear. Springer, New York.
- Kim, T. S., T. Nakagawa, J. E. Lee, K. Fujino, F. Iguchi et al., 2004. Induction of cell proliferation and beta-catenin expression in rat utricles in vitro. Acta Otolaryngol. Suppl. 551: 22–25. [DOI] [PubMed] [Google Scholar]
- Lebel, M., P. Agarwal, C. W. Cheng, M. G. Kabir, T. Y. Chan et al., 2003. The Iroquois homeobox gene Irx2 is not essential for normal development of the heart and midbrain-hindbrain boundary in mice. Mol. Cell. Biol. 23: 8216–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. S., F. Liu and N. Segil, 2006. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development 133: 2817–2826. [DOI] [PubMed] [Google Scholar]
- Leon, Y., S. Sanchez-Galiano and I. Gorospe, 2004. Programmed cell death in the development of the vertebrate inner ear. Apoptosis 9: 255–264. [DOI] [PubMed] [Google Scholar]
- Lin, J., M. Ozeki, E. Javel, Z. Zhao, W. Pan et al., 2003. Identification of gene expression profiles in rat ears with cDNA microarrays. Hear. Res. 175: 2–13. [DOI] [PubMed] [Google Scholar]
- Liu, X., J. A. Mohamed and R. Ruan, 2004. Analysis of differential gene expression in the cochlea and kidney of mouse by cDNA microarrays. Hear. Res. 197: 35–43. [DOI] [PubMed] [Google Scholar]
- Lumpkin, E. A., T. Collisson, P. Parab, A. Omer-Abdalla, A. Haeberle et al., 2003. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3: 389–395. [DOI] [PubMed] [Google Scholar]
- Marazita, M. L., L. M. Ploughman, B. Rawlings, E. Remington, K. S. Arnos et al., 1993. Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. Am. J. Med. Genet. 46: 486–491. [DOI] [PubMed] [Google Scholar]
- Mclaughlin, L., G. Zhu, M. Mistry, C. Ley-Ebert, W. D. Stuart et al., 2000. Apolipoprotein J/clusterin limits the severity of murine autoimmune myocarditis. J. Clin. Invest. 106: 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehl, A. L, and V. Thompson, 1998. Newborn hearing screening: the great omission. Pediatrics 101: e4. [DOI] [PubMed] [Google Scholar]
- Morsli, H., D. Choo, A. Ryan, R. Johnson and D. K. Wu, 1998. Development of the mouse inner ear and origin of its sensory organs. J. Neurosci. 18: 3327–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino, J., K. Yamashita, H. Hashiguchi, H. Fujii, T. Shimazaki et al., 2004. Meteorin: a secreted protein that regulates glial cell differentiation and promotes axonal extension. EMBO J. 9: 1998–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niskar, A. S., S. M. Kieszak, A. Holmes, A. Esteban, C. Rubin et al., 1998. Prevalence of hearing loss among children 6 to 19 years of age: the third national health and nutrition examination survey. J. Am. Med. Assoc. 279: 1071–1075. [DOI] [PubMed] [Google Scholar]
- Ohyama, T., O. A. Mohamed, M. M. Taketo, D. Dufort and A. K. Groves, 2006. Wnt signals mediate a fate decision between otic placode and epidermis. Development 133: 865–875. [DOI] [PubMed] [Google Scholar]
- Oishi, K., K. Miyazaki, K. Kadota, R. Kikuno, T. Nagase et al., 2003. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J. Biol. Chem. 278: 41519–41527. [DOI] [PubMed] [Google Scholar]
- Parving, A., 1993. Congenital hearing disability, epidemiology and identification: A comparison between two health authority districts. Int. J. Pediatr. Otorhinolaryngol. 27: 29–46. [DOI] [PubMed] [Google Scholar]
- Pauley, S., E. Lai and B. Fritzsch, 2006. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev. Dyn. 235: 2470–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola, U., J. Ylikoski, R. Trokovic, J. M. Hebert, S. K. Mcconnell et al., 2002. FGFR1 is required for the development of the auditory sensory epithelium. Neuron 35: 671–680. [DOI] [PubMed] [Google Scholar]
- Raphael, Y., and R. A. Altschuler, 2003. Structure and innervation of the cochlea. Brain Res. Bull. 60: 397–422. [DOI] [PubMed] [Google Scholar]
- Reich, M., K. Ohm, P. Tamayo, M. Angelo and J. P. Mesirov, 2004. GeneCluster 2.0: an advanced toolset for bioarray analysis. Bioinformatics 20: 1797–1798. [DOI] [PubMed] [Google Scholar]
- Resendes, B. L., N. G. Robertson, J. D. Szustakowski, R. J. Resendes, Z. Weng et al., 2002. Gene discovery in the auditory system: characterization of additional cochlear-expressed sequences. J. Assoc. Res. Otolaryngol. 3: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, N. G., U. Khetarpal, G. A. Gutierrez-Espelata, F. R. Bieber and C. C. Morton, 1994. Isolation of novel and known genes from a human fetal cochlear cDNA library using subtractive hybridization and differential screening. Genomics 23: 42–50. [DOI] [PubMed] [Google Scholar]
- Ruben, R. J., 1967. Development of the inner ear of the mouse: a radioautographic study of terminal mitosis. Acta Otolaryngol. 220: 1–44. [PubMed] [Google Scholar]
- Santilli, G., B. J. Aronow and A. Sala, 2003. Essential requirement of apolipoprotein J (clusterin) signaling for IkappaB expression and regulation of NF-kappaB activity. J. Biol. Chem. 278: 38214–38219. [DOI] [PubMed] [Google Scholar]
- Shailam, R., P. J. Lanford, C. M. Dolinsky, C. R. Norton, T. Gridley et al., 1999. Expression of proneural and neurogenic genes in the embryonic mammalian vestibular system. J. Neurocytol. 28: 809–819. [DOI] [PubMed] [Google Scholar]
- Shi, C., X. Zhang, Z. Chen, K. Sulaiman, M. W. Feinberg et al., 2004. Integrin engagement regulates monocyte differentiation through the forkhead transcription factor Foxp1. J. Clin. Invest. 114: 408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, S., and S. A. Fuqua, 2001. Estrogen receptor and breast cancer. Semin. Cancer Biol. 11: 339–352. [DOI] [PubMed] [Google Scholar]
- Stenberg, A. E., H. Wang, L. Sahlin and M. Hultcrantz, 1999. Mapping of estrogen receptors alpha and beta in the inner ear of mouse and rat. Hear. Res. 136: 29–34. [DOI] [PubMed] [Google Scholar]
- Stenberg, A. E., H. Wang, L. Sahlin, P. Stierna, E. Enmark et al., 2002. Estrogen receptors alpha and beta in the inner ear of the ‘Turner mouse’ and an estrogen receptor beta knockout mouse. Hear. Res. 166: 1–8. [DOI] [PubMed] [Google Scholar]
- Stevens, C. B., A. L. Davies, S. Battista, J. H. Lewis and D. M. Fekete, 2003. Forced activation of Wnt signaling alters morphogenesis and sensory organ identity in the chicken inner ear. Dev. Biol. 261: 149–164. [DOI] [PubMed] [Google Scholar]
- Sulik, K. K., 1995. Embryology of the ear, pp 22–42 in Hereditary Hearing Loss and Its Syndromes, Ed. 1, edited by R. J. Gorlin, H. V. Toriello and M. M. Cohen. Oxford University Press, New York.
- Takebayashi, S., T. Nakagawa, K. Kojima, T. S. Kim, T. Kita et al., 2004. Expression of beta-catenin in developing auditory epithelia of mice. Acta Otolaryngol. Suppl. 551: 18–21. [DOI] [PubMed] [Google Scholar]
- Takeuchi, S., M. Ando and A. Kakigi, 2000. Mechanism generating endocochlear potential: Role played by intermediate cells in stria vascularis. Biophys. J. 79: 2572–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo, P., D. Slonim, J. Mesirov, Q. Zhu, E. Dmitrovsky et al., 1999. Interpreting gene expression with self-organizing maps: methods and application to hematopoeitic differentiation. Proc. Natl. Acad. Sci. USA 96: 2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos, A. C., J. F. Berson, S. C. Theos, K. E. Herman, D. C. Harper et al., 2006. Dual loss of ER export and endocytic signals with altered melanosome morphology in the silver mutation of Pmel17. Mol. Biol. Cell. 8: 3598–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama, K., M. Ozeki, Y. Hamajima and J. Lin, 2005. Expression of the integrin genes in the developing cochlea of rats. Hear. Res. 201: 21–26. [DOI] [PubMed] [Google Scholar]
- Tsinkalovsky, O., E. Filipski, B. Rosenlund, R. B. Sothern, H. G. Eiken et al., 2006. Circadian expression of clock genes in purified hematopoietic stem cells is developmentally regulated in mouse bone marrow. Exp. Hematol. 34: 1249–1261. [DOI] [PubMed] [Google Scholar]
- Tusher, V., R. Tibshirani and G. Chu, 2001. Significance analysis of microarrays applied to transcriptional responses to ionizing radiation. Proc. Natl. Acad. Sci. USA 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpy, E., M. Leibovici and C. Petit, 1999. Characterization of otoconin-95, the major protein of murine otoconia, provides insights into the formation of these inner ear biominerals. Proc. Natl. Acad. Sci. USA. 96: 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., J. Weidenfeld, M. M. Lu, S. Maika, W. A. Kuziel et al., 2004. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development 131: 4477–4487. [DOI] [PubMed] [Google Scholar]
- Warchol, M. E., 1997. Macrophage activity in organ cultures of the avian cochlea: demonstration of a resident population and recruitment to sites of hair cell lesions. J. Neurobiol. 33: 724–734. [PubMed] [Google Scholar]
- Warchol, M. E., 1999. Immune cytokines and dexamathasone influence sensory regeneration in the avian vestibular periphery. J. Neurocytol. 28: 889–900. [DOI] [PubMed] [Google Scholar]
- Warchol, M. E., and B. A. Kaplan, 1999. Macrophage secretory products influence the survival of statoacoustic neurons. Neuroreport 10: 665–668. [DOI] [PubMed] [Google Scholar]
- Warchol, M. E., J. I. Matsui, E. L. Simkus and J. M. Ogilive, 2001. Ongoing cell death and immune influences on regeneration in the vestibular sensory organs. Ann. NY Acad. Sci. 942: 34–45. [DOI] [PubMed] [Google Scholar]
- Wilson, M. R., and S. B. Easterbrook-Smith, 2000. Clusterin is a secreted mammalian chaperone. Trends Biochem. Sci. 25: 95–98. [DOI] [PubMed] [Google Scholar]
- Yamamoto, T., Y. Nakahata, H. Soma, M. Akashi, T. Mamine et al., 2004. Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Mol. Biol. 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J. L., J. Shou, F. Guillemot, R. Kageyama and W-Q. Gao, 2000. Hes1 is a negative regulator of inner ear hair cell differentiation. Development 127: 4551–4560. [DOI] [PubMed] [Google Scholar]
- Zine, A., A. Aubert, J. Qiu, S. Therianos, F. Guillemot et al., 2001. Hes1 and Hes5 activities are required for the normal development of the hair cells in the mammalian inner ear. J. Neurosci. 21: 4712–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]