Abstract
We describe a second-generation deficiency kit for Drosophila melanogaster composed of molecularly mapped deletions on an isogenic background, covering ∼77% of the Release 5.1 genome. Using a previously reported collection of FRT-bearing P-element insertions, we have generated 655 new deletions and verified a set of 209 deletion-bearing fly stocks. In addition to deletions, we demonstrate how the P elements may also be used to generate a set of custom inversions and duplications, particularly useful for balancing difficult regions of the genome carrying haplo-insufficient loci. We describe a simple computational resource that facilitates selection of appropriate elements for generating custom deletions. Finally, we provide a computational resource that facilitates selection of other mapped FRT-bearing elements that, when combined with the DrosDel collection, can theoretically generate over half a million precisely mapped deletions.
THE availability of chromosomal deletion collections is of considerable benefit to the Drosophila research community for gene mapping, the phenotypic characterization of alleles, and genomewide genetic interaction screens. A core deficiency kit, composed of 270 genetically heterogeneous deletions covering ∼92% of the genome, has been built up over many years by the Bloomington Drosophila Stock Center (BDSC; http://flystocks.bio.indiana.edu/Browse/df-dp/dfkit-info.htm). Continuing efforts by the Bloomington Center are currently focused on expanding genome coverage by recovering deletions in the vicinity of haplo-insufficient regions (K. Cook, personal communication). Despite the considerable utility of this collection, it does, by its very nature, suffer from a number of limitations. These include a heterogeneous genetic background, the presence of uncharacterized second-site mutations, and, for most deletions, molecularly undefined breakpoints. More recently, two groups have taken advantage of two key technologies: large collections of transposon insertions precisely mapped to the Drosophila genome sequence and site-specific recombination, to develop tools for producing custom chromosomal deletions in homogeneous genetic backgrounds that are mapped to the genome sequence with single-base-pair resolution (Parks et al. 2004; Ryder et al. 2004; Thibault et al. 2004).
In both cases, the new deletion collections are generated using FLP-mediated recombination between pairs of transposon-borne FRT sites, a method originally developed in Drosophila by Golic and Golic (1996). In one case (Parks et al. 2004), a set of >29,000 P-element and piggyBac insertions (Thibault et al. 2004) were used to generate 519 deletions covering 56% of the euchromatic genome (the Exelixis collection). The high number of starting insertions used by this group allows fine-scale coverage of the genome with relatively small deletions; the average size of the existing collection is ∼140 kb and facilitates the ongoing efforts of BDSC to increase genome coverage. While this collection provides a route for mapping and screening particular regions of the genome at a relatively high resolution, the fact that >1000 deletions of this size are needed to cover the genome makes it less suitable for high-throughput genomewide screens; with 270 stocks, the traditional deficiency kit is more useful in this respect. In constructing our deficiency collection we have taken a similar approach to Parks et al. (2004); however, we generated deletions with a larger average size and thus provide a complementary resource to their collection. Thus genomewide screens in defined genetic backgrounds can be rapidly performed at medium resolution using the DrosDel collection, and, subsequently, specific regions can be targeted at higher resolution using Exelixis or BDSC deletions.
In this article, we describe the expansion of the DrosDel P-element collection and its use in constructing a genomewide deletion set, covering ∼77% of the euchromatic genome on a single isogenic genetic background. As described by Golic and Golic (1996), recombination between FRT sites can be used to create other precisely mapped chromosomal aberrations such as inversions and duplications. Using our insert collection, we present methods for constructing deletions in “difficult” regions of the genome, for example, those harboring haplo-insufficient loci, by generating covering duplications. These methods complement the approaches being taken by BDSC and hold out the prospect of generating complete deletion coverage of the Drosophila melanogaster genome. Finally, we describe how FRT-bearing elements from the DrosDel and Exelixis collections can be combined to generate a theoretical set of >500,000 precisely mapped deletions, and we introduce a simple computational interface for mining these FRT-derived deletions (FDDs).
MATERIALS AND METHODS
Mapping of P elements:
Mapping of elements for the collection was performed by inverse PCR and sequencing as described previously (Ryder et al. 2004). Along with new insertions, all existing mapped elements were realigned to Release 5.1 of the Drosophila genome. Additional information can be obtained from the DrosDel website (http://www.drosdel.org.uk).
Construction of chromosomal aberrations:
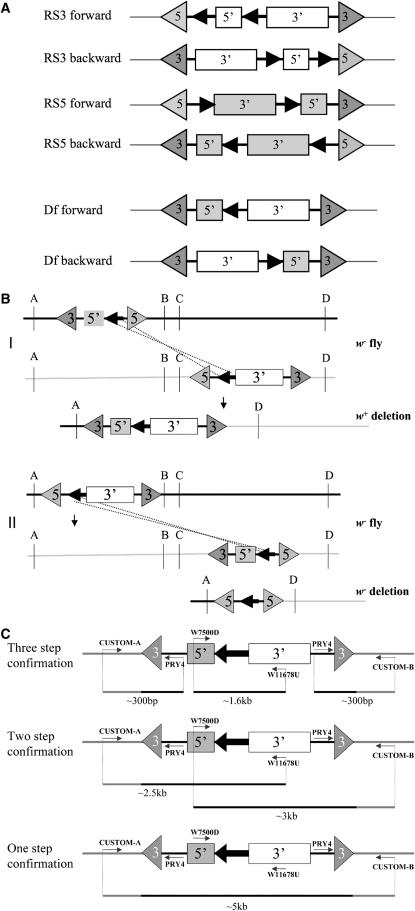
The structure of the RS3 and RS5 constructs means that the FRT sites and fragments of the white gene are in different orientations depending on the element type (Figure 1A). This must be taken into consideration when designing aberrations and Figure 1B shows the outcomes of recombination events between elements in different relative orientations with respect to the chromosome. The orientation of elements is based primarily on the P-element ends in relation to the genome scaffold (for example, an element in the forward orientation would be 5′ ← 5′P = 3′P → 3′) and we refer to this orientation as P(F) and its inverse as P(R). Due to the structure of the RS elements, the orientation of the internal FRT sites differs, depending on the element type. When referring explicitly to the FRT orientation, the terms F(F) or F(R) will be used. We have designated inversions created by FLP-mediated recombination using RS elements as EIN [European inversions; e.g., In(2L)EIN1] and duplications generated by recombining these inversions as EDP [European duplications; e.g., Dp(2;2)EDP1]. New deletions generated by recombining inversions are named after their inverted progenitors and, since they are inversions, are given an EIN designation [e.g., In(2L)EIN17L EIN30R]; see Table 3.
Figure 1.—
Structure of RS elements. (A) The orientation of RS3 and RS5 elements and their recombination products. 5′ and 3′ exons of the mini-white are marked by the open (RS3) or shaded (RS5) boxes. P-element ends are indicated by the triangles. FRT sites are designated by the solid arrows. (B) Recombination between an RS3r and an RS5r element in trans can generate a w+ deletion (I) or a w deletion (II), depending upon the relative orientations of the elements with respect to the chromosome. (C) Molecular strategies for confirming deletions (see materials and methods for details). The RS elements are as described in A. The locations of the primers described in materials and methods are shown as thin arrows with size of the PCR product indicated below each product.
TABLE 3.
2L inversions
| Inversion | RS element 1 | Scaffold 1 | RS element 1 | Scaffold 2 |
|---|---|---|---|---|
| In(2L)EIN1 | 5-HA-1160 | 4701129 | CB-0304-3 | 19158440 |
| In(2L)EIN2 | 5-HA-1191 | 67365 | CB-0304-3 | 19158440 |
| In(2L)EIN3 | 5-HA-1535 | 2753125 | CB-0304-3 | 19158440 |
| In(2L)EIN4 | 5-HA-1621 | 4892305 | CB-0304-3 | 19158440 |
| In(2L)EIN5 | 5-HA-1706 | 9437469 | CB-0304-3 | 19158440 |
| In(2L)EIN6 | 5-HA-1707 | 4452979 | CB-0304-3 | 19158440 |
| In(2L)EIN7 | 5-HA-1711 | 5980272 | CB-0304-3 | 19158440 |
| In(2L)EIN8 | 5-HA-1712 | 6268819 | CB-0304-3 | 19158440 |
| In(2L)EIN9 | 5-HA-1999 | 7010116 | CB-0304-3 | 19158440 |
| In(2L)EIN10 | 5-HA-2004 | 3055770 | CB-0304-3 | 19158440 |
| In(2L)EIN11 | 5-HA-2414 | 2299231 | CB-0304-3 | 19158440 |
| In(2L)EIN12 | 5-HA-3051 | 5055158 | CB-0304-3 | 19158440 |
| In(2L)EIN13 | 5-HA-5091 | 3632183 | CB-0304-3 | 19158440 |
| In(2L)EIN14 | 5-SZ-3127 | 8205159 | CB-0304-3 | 19158440 |
| In(2L)EIN15 | 5-SZ-3139 | 9205076 | CB-0304-3 | 19158440 |
| In(2L)EIN16 | 5-SZ-3337 | 6709099 | CB-0304-3 | 19158440 |
| In(2L)EIN17 | 5-SZ-3596 | 568095 | CB-0304-3 | 19158440 |
| In(2L)EIN18 | 5-SZ-3622 | 8415721 | CB-0304-3 | 19158440 |
| In(2L)EIN19 | 5-SZ-3985 | 6000124 | CB-0304-3 | 19158440 |
| In(2L)EIN20 | 5-SZ-3989 | 1737465 | CB-0304-3 | 19158440 |
| In(2L)EIN21 | 5-SZ-4117 | 5801918 | CB-0304-3 | 19158440 |
| In(2L)EIN22 | CB-0110-3 | 4892105 | 5-HA-1724 | 18823590 |
| In(2L)EIN23 | CB-0114-3 | 3018404 | 5-HA-1724 | 18823590 |
| In(2L)EIN24 | CB-0279-3 | 10732704 | 5-HA-1724 | 18823590 |
| In(2L)EIN25 | CB-0473-3 | 9581740 | 5-HA-1724 | 18823590 |
| In(2L)EIN26 | CB-0536-3 | 6963808 | 5-HA-1724 | 18823590 |
| In(2L)EIN27 | CB-0621-3 | 5027473 | 5-HA-1724 | 18823590 |
| In(2L)EIN28 | CB-0716-3 | 5949427 | 5-HA-1724 | 18823590 |
| In(2L)EIN29 | CB-0886-3 | 6648731 | 5-HA-1724 | 18823590 |
| In(2L)EIN30 | CB-5353-3 | 587983 | 5-HA-1724 | 18823590 |
| In(2L)EIN31 | CB-5692-3 | 2197121 | 5-HA-1724 | 18823590 |
| In(2L)EIN32 | CB-6167-3 | 8205470 | 5-HA-1724 | 18823590 |
| In(2L)EIN33 | CB-6222-3 | 5237390 | 5-HA-1724 | 18823590 |
| In(2L)EIN34 | UM-8100-3 | 7576637 | 5-HA-1724 | 18823590 |
| In(2L)EIN35 | UM-8380-3 | 5659293 | 5-HA-1724 | 18823590 |
| In(2L)EIN36 | 5-HA-1693 | 183037 | CB-6227-3 | 19791763 |
| In(2L)EIN37 | 5-HA-1693 | 183037 | CB-0898-3 | 19158447 |
| In(2L)EIN38 | 5-HA-1693 | 183037 | CB-0522-3 | 13717341 |
| In(2L)EIN39 | 5-HA-1693 | 183037 | UM-8369-3 | 12436439 |
| In(2L)EIN40 | 5-SZ-3548 | 207391 | CB-5235-3 | 18151698 |
| In(2L)EIN41 | 5-SZ-3548 | 207391 | CB-5425-3 | 16281817 |
| In(2L)EIN42 | 5-SZ-3548 | 207391 | CB-5697-3 | 15061074 |
| In(2L)EIN43 | 5-SZ-3548 | 207391 | CB-5032-3 | 13878181 |
| In(2L)EIN44 | 5-SZ-3548 | 207391 | CB-0787-3 | 12055953 |
| In(2L)EIN45 | 5-SZ-3548 | 207391 | CB-5014-3 | 12045104 |
| In(2L)EIN46 | 5-SZ-3548 | 207391 | UM-8151-3 | 10474364 |
| In(2L)EIN47 | 5-SZ-3548 | 207391 | CB-0304-3 | 19158440 |
| In(2L)EIN48 | 5-HA-1614 | 249337 | CB-0304-3 | 19158440 |
Inversions were generated in the distal half of 2L by recombination between the listed elements. The scaffold locations with respect to the Release 5.1 sequence are given. See text for details.
Deletions:
Deletion crosses were performed as described previously (Ryder et al. 2004; http://www.drosdel.org.uk). A computer program was designed to select pairs of RS3 and RS5 elements that were <1 Mb apart and in the correct orientation relative to the chromosome and to each other. Fly stocks carrying RS elements of interest were heat-shocked in the presence of 70FLP to remove part of the mini-white gene, and the resulting reduced RSr elements were isolated as white-eyed progeny. Flies carrying both the two reduced elements in trans and 70FLP were heat-shocked to construct the deletions and subsequently isolated as exceptional w+ progeny.
Tip deletions:
The method used for construction of tip deletions was identical to that for normal intrachromosomal deletions (Ryder et al. 2004) except that the two starting elements selected were very close to the ends of two chromosomes, one 19 kb from the tip of the X and the others ∼100 kb from the tips of both arms of chromosomes 2 and 3. The resulting deletions are nonreciprocal translocations in which autosomal terminal deletions are capped with the tip of the X.
Inversions:
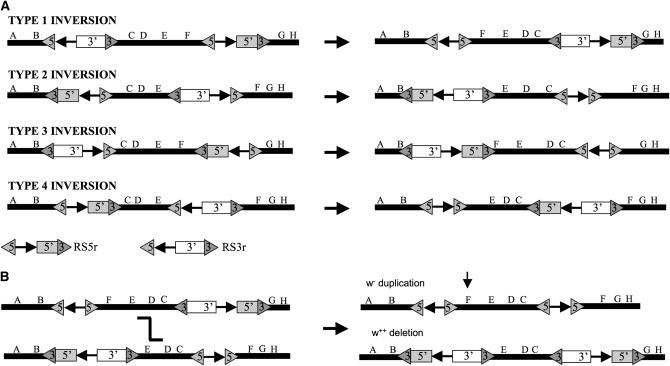
Inversions were constructed by the FLP–FRT method from RS3 and RS5 elements carried in cis and in the same orientation (Golic and Golic 1996). One of the resulting breakpoints of these inversions carries a reconstituted w+ and an FRT site; the other breakpoint carries a single FRT site with no associated w gene. Several types of inversion can be constructed and are designated types 1–4 (Figure 2A). Which breakpoint carries w+ is determined by the orientation of the FRT sites.
Figure 2.—
Inversion types and the generation of aneuploid chromosomes. (A) Four types of inversion (right) are possible, depending upon the structure of the parental chromosome carrying the RS elements in cis (left). For clarity, we show the reduced forms of the elements (RSr). (B) Recombination between pairs of inversions produces aneuploid chromosomes. For illustration, we show the products of an exchange between a type 1 and a type 2 inversion, a w duplication (the parental line is w+) and a deletion carrying two copies of w+. Since the nonrecombinant progeny carry only a single copy of w+, the deletion may be identifiable by virtue of a darker eye color.
Duplications and deletions derived from inversions:
Duplications (and deletions) may be isolated as a result of exchange between two similar inversions (Figure 2B) (Muller 1930). Recombination within the inverted region results in aneuploidy for the regions between the inversions' breakpoints. Pairs of inversions were selected with one similar breakpoint and one breakpoint differing by up to 2.9 Mb. Crossing over between the inversions resulted in duplications of these regions, which could then be used to recover deletions that would otherwise be haplo-lethal or haplo-sterile. Where possible, inversions carrying the w+ marker on opposite breakpoints were selected, allowing the duplications to be isolated as the only w progeny. Note that the reciprocal recombinants are deletions and carry a w+ marker on each of the breakpoints. In most combinations the deletion chromosome can be identified by the additive phenotypic effect of the two w+ genes. Alternatively, duplications of some regions may be isolated by suppression of the phenotype of a haplo-insufficient or antimorphic mutation, mapping to the region of interest [e.g., a Minute or Su(var)]. These duplications are “nested” within inversions and are therefore stable. It has been demonstrated, particularly in the case of Dp(1;1)B, that tandem duplications are unstable with loss of the duplication occurring by unequal exchange in duplication homozygotes (Sturtevant 1925; Tsubota 1991) and, less frequently, by sister-chromatid exchange (Peterson and Laughnan 1963).
Confirmation of deletions:
Deletions were confirmed using both molecular and genetic methods. For genetic confirmation, putative deficiency lines were crossed with stocks that carry a molecularly defined visible or lethal mutation predicted to be uncovered by the deletion. Failure of a putative deficiency stock to complement these mutations strongly suggests that the deletion is present.
Figure 1C shows three potential methods for confirming deletions at the molecular level. As a tool to aid deletion confirmation, primers were automatically designed for all predicted deletions in the DrosDel collection using a Perl script linked to Primer3 (Rozen and Skaletsky 2000). Primer3 parameters (minimum anneal, 50°; maximum primer length, 26; minimum CG%, 18) were chosen to pick primers ∼300 bp away from the P-element ends. As the three-step process was used routinely in the laboratory, the primers were paired with the PRY4 primer (CAATCATATCGCTGTCTCACTCA) for design purposes; however, in most cases they should also work in combination for the one-step confirmation protocol. The presence of the reconstituted w gene in the three-step process was determined by amplification across the FRT site using the W7500D (GTCCGCCTTCAGTTGCACTT) and W11678U (TCATCGCAGATCAGAAGCGG) primers as originally described by Golic and Golic (1996). For the one-step and two-step confirmation, long-range PCR was performed using the custom primers designed for the three-step confirmation and the Expand long template PCR system (Roche Diagnostics) using the standard “system 1” (two-step) or “system 2” (one-step) protocol.
For conventional polytene chromosome analysis, we used propionic carmine–orcein squash preparations (Ashburner 1989). In situ hybridizations were performed with biotinylated probes and horseradish peroxidase detection according to standard protocols (Ashburner 1989). Polytene chromosomes were interpreted using the revised maps of C. B. Bridges and P. N. Bridges (see Lefevre 1976).
RESULTS
Update of P-element collection:
The current number of mapped RS elements that we have processed is shown in Table 1. After eliminating 109 lines according to our previously described criteria (Ryder et al. 2004), a total of 3332 elements remain, adding a further 89 insertions to our collection. Each of these RS insertions maps to an unambiguous location on the Release 5.1 genome sequence and full details for each insertion, along with a collection of search tools, are available via the DrosDel website. These sequence data have been submitted to GenBank (accession nos. AJ545047–AJ547612 and AJ622065–AJ622812) and are also incorporated in FlyBase (http://www.flybase.org).
TABLE 1.
Summary of the DrosDel RS collection and deletions constructed
| Chromosome arm | No. of mapped RS inserts per chromosome arm | No. of deletions constructed | No. of deletions molecularly confirmed by PCR | No. of deletions confirmed by complementation | No. of deletions confirmed by both molecular and genetic tests | Average deletion size for each chromosome arm (kb) | Average no. of genes removed by deletion for each chromosome arm |
|---|---|---|---|---|---|---|---|
| 2L | 659 | 276 | 137 | 22 | 15 | 328 | 38 |
| 2R | 736 | 74 | 41 | 39 | 33 | 459 | 62 |
| 3L | 538 | 67 | 48 | 55 | 45 | 475 | 54 |
| 3R | 696 | 169 | 85 | 104 | 82 | 376 | 46 |
| 4 | 23 | 8 | 6 | 8 | 6 | 313 | 19 |
| X | 680 | 71 | 53 | 11 | 4 | 311 | 33 |
| Total | 3332 | 665 | 370 | 239 | 189 | 377 | 42 |
Theoretical and computational generation of deletions:
Deletions were designed on the basis that the maximum deficiency that a fly can reasonably tolerate is ∼1 Mb (Ashburner et al. 2005). Since the mapping of the original elements will produce different strand matches, depending on which end was amplified or element type used, a script was first used to orientate the elements in relation to their P-element ends [P(F) and P(R)]. For each element in a given orientation, the data set was scanned for elements of a different type that were within 1 Mb and in an orientation that would produce a functional reconstituted white gene after FRT-mediated recombination. The correct relative orientation of the elements with respect to each other (Figure 1B) is important if deletions are to be selected on the basis of eye color. The correct orientation of elements produces a deletion with a w+ phenotype (with a reciprocal w duplication), whereas other orientations produce a w+ duplication and a phenotypically untraceable w deletion. Note, however, that w deletions may be selected via a molecular screen, for example, using a sib-selection strategy (Kaiser and Goodwin 1990). The script outputs a text-delimited table that is imported into MySQL for further manipulation and querying.
Generation of DrosDel kit:
Using the DrosDel collection of RS3 and RS5 insertions, nearly 13,000 w+ deletions between 1 bp and 1 Mb in length can theoretically be constructed, making the collection a powerful resource for researchers wanting to generate custom deletions in regions of interest (Table 2). Since our aim is to generate a second-generation “deletion kit,” we sought to cover as much of the genome as possible with mapped deficiencies. To do this, we chose potential deletions of <1 Mb and from these selected a tiling path of deletions that would cover the euchromatic genome with as few stocks as possible. We were careful to avoid known haplo-insufficient loci (S. Marygold and K. Cook, personal communication).
TABLE 2.
Coverage statistics for the DrosDel deletion collection
| Chromosome arm | Chromosome length (bp) | No. of base pairs of each chromosome arm covered by made DrosDel deletions (bp) | No. of base pairs of each chromosome arm covered by the core DrosDel kit (bp) | Possible coverage: theoretical coverage of each chromosome by DrosDel deletions (bp) | Possible deletions: the theoretical no. of deletions that can be constructed on each chromosome arm |
|---|---|---|---|---|---|
| 2L | 23,011,544 | 20,363,603 (88.49) | 15,333,203 (66.63) | 21,617,081 (93.94) | 2,457 |
| 2R | 21,146,708 | 16,909,704 (79.96) | 10,902,028 (51.55) | 20,266,146 (95.84) | 4,034 |
| 3L | 24,543,557 | 17,476,562 (71.21) | 15,279,214 (62.25) | 23,591,187 (96.12) | 1,528 |
| 3R | 27,905,053 | 23,260,221 (83.35) | 19,228,295 (68.91) | 27,843,696 (99.78) | 2,158 |
| 4 | 1,351,857 | 849,757 (62.86) | 849,757 (62.86) | 849,757 (62.86) | 26 |
| X | 22,422,827 | 14,110,967 (62.93) | 11,218,881 (50.03) | 22,263,078 (99.29) | 2,762 |
| Total | 120,381,546 | 92,970,814 (77.23) | 72,811,378 (60.48) | 116,430,945 (96.72) | 12,965 |
Numbers in parentheses are percentages. All data are with respect to Release 5.1 of the genome sequence.
To generate the deletion stocks, we took a consortium approach, where several laboratories each focused on a particular region of the genome. The genome was split into its component chromosome arms (or sections of arms for the larger chromosomes) and each group in the consortium concentrated efforts on its designated section (X: Zurich and Cambridge; 2L: Halle; 2R: Umea and Barcelona; 3L: Szeged; 3R: Heidelberg, Mainz, and Halle; 4: Cambridge). To date, 870 different deletions have been attempted by the consortium and, of these, 665 (76%) were recovered—a very high success rate. We were unable to stabilize all deletions as balanced stocks, however. Several were dominant male and/or female sterile, lethal, or sterile over balancers or generally too sickly to be maintained without constant attention. In some cases, this lack of viability could be attributed to particular haplo-insufficient loci or, in the case of large deletions, presumably to the additive deleterious effects of haploidy for many genes (data not shown). A summary and detailed statistics on the collection are provided in Tables 1 and 2 and the list of all deletions is provided in supplemental Table 1 at http://www.genetics.org/supplemental/.
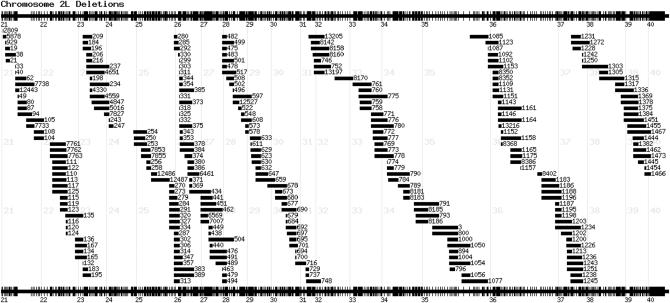
Although the original concept was to create minimal overlap coverage with as few deletions as possible, in practice this protocol was changed in light of several practical issues. These included failed deletions, the paucity of elements in some regions, and the detection of false-positive lines. In addition, we found that some stocks are too sick to be kept in the long term or proved to be impossible to balance, making them inappropriate for stock center maintenance. To this end, we determined a new “core kit” of deletions, which have been confirmed by PCR and/or by genetic analysis where possible: these are available to the community via the Szeged and Bloomington stock centers. Maps of the deletion tiling paths are available from the DrosDel website (http://www.drosdel.org.uk/coverage.php). This core kit of verified deletions comprises 209 stocks and is predicted to cover 60% of the euchromatic genome (Table 2; supplemental Table 2 at http://www.genetics.org/supplemental/). The coverage of all deletions that have been constructed by the DrosDel consortium is >77% of the genome (Figure 3 shows an example of coverage on chromosome 2L). While not all these are currently available from stock centers due to balancing issues and stock health, they may be requested from individual labs via the Szeged stock center. Theoretically, the RS elements in the DrosDel kit are capable of covering nearly 97% of the euchromatic genome. However, in practice, this coverage level cannot be achieved using simple deletions due to haplo-insufficient regions.
Figure 3.—
Map of the deletion coverage for chromosome arm 2L. The cytological map of 2L is given at the top and bottom, with the extent of each of the DrosDel deletions indicated. For clarity, the Df(2)ED prefix is omitted and only the deletion numbers are given.
Confirmation of deletions:
Three methods were used for molecularly confirming deletions via PCR (Figure 1C). The three-step process amplifies the 3′-ends of both parental elements and separately confirms that the reconstituted white gene is present. It does not, however, unambiguously confirm that the white gene is associated with the P element or that a deletion is present. Neither the two-step process, amplifying from both ends across the FRT, nor the one-step process, amplifying across the entire RS5+3 element, could be used routinely due to the difficulties encountered when attempting to consistently amplify large PCR products. For this reason, we confirmed the deletions by the three-step method, and subsequently reconfirmed by genetic complementation where possible. We strongly recommend, however, that groups who create their own deletions with the DrosDel system attempt to use the one-step confirmation method. A subset of deletions was also confirmed by cytological analysis of polytene chromosomes (Figure 4).
Figure 4.—
Cytological verification of DrosDel deletions. Each deletion is heterozygous with a wild-type chromosome and the arrows indicate the location of the deletion. (A) Df(3R)ED6316 (99A5; 99C1, 527 kb). (B) Df(3L)ED4177 (61C2; 61E2, 715 kb). (C) Df(3L)ED4475 (68C13; 69B4, 821 kb).
The results of the deletion construction efforts are summarized in Tables 1 and 2. Of the 665 putative deletions that we have constructed, 56% (370) have been molecularly confirmed by PCR. Seventeen percent (111) gave an ambiguous result in one of the three-step PCR assays and could not be confirmed in this manner; this does not necessarily indicate that these deletions are false positives, but they should be viewed with a degree of caution. These chromosomes should be tested by the one-step PCR method. We note that, since the strategy requires reconstitution of a functional w gene, then imprecise breakpoints due to chromosome resection will not be recovered since such events would eliminate w before removing flanking genomic DNA. The remaining 27% (184) of the collection has not yet been tested. In addition to molecular analysis, 36% (239) of the deletions were confirmed when assessed by genetic complementation, assaying whether they uncovered a molecularly mapped mutation. Although a single complementation test cannot assess the extent of a deletion or confirm the precision of the breakpoints, we have carefully tested several deletions in the Adh region, where we have extensive genetic data, in some cases against mutations of adjacent genes, to identify any potential problems. For example, Df(2L)ED3 is predicted to partially delete noc, and ED3/noc4 does indeed have a weak noc phenotype. The proximal breakpoints of Df(2L)ED3, Df(2L)ED800, and Df(2L)ED1000 are within 1 kb of each other and are predicted to be between nht and esg. All three deletions do indeed delete nht (ED/nhtz5347 are male sterile), but not the adjacent esg (ED/esg35Ce-1 are viable). An additional six deletion breakpoints were tested by precise genetic assays; all behaved as expected and support the view that this method of deletion construction is accurate.
A total of 28% of the collection (189 deletions) has been confirmed both by a molecular assay and by a complementation test. A total of 23 putative deletions failed the genetic tests and were therefore classed as false positives and discarded. Note that we focused on confirmation of the core kit in the first instance and are gradually confirming the remainder. Of the 209 deletions in the core collection, 89% have been confirmed by PCR, 64% confirmed by genetic complementation, and 54% by both methods. Data on molecular and genetic confirmation are provided for each deletion at the DrosDel website.
There are several reasons why we may not recover particular deletions. In the most trivial cases, especially for larger deletions, we may simply have failed to screen enough progeny and it is possible that a given deletion may be recovered in a larger-scale cross. It is also possible that some of the failed deletions were not recovered because they uncover unmapped haplo-insufficient regions. We encountered a variable level of false-positive recovery, depending upon the deletion being attempted. In the majority of crosses, all the progeny were of the expected genotype; however, ∼6% of the deletion crosses segregated red-eyed individuals that produced homozygous viable lines or lines that failed genetic complementation tests or lines in which the w+ mapped to the wrong chromosome. These are unlikely to be carrying deletions and were discarded as false positives. Our current view is that false positives result from aberrant recombination events mediated by the FLP recombinase, but we have not investigated the nature of these chromosomes further.
Homozygous viable w+ lines were not always false positives. For example, we found that when making Df(1)ED7635, a 278-kb deletion in region 19B, viable w+ males were produced although they were weak and sterile. Since we expected the deletion-carrying males to be nonviable, we presumed that the cross was generating a false positive. We were surprised to find that the deletion was confirmed by PCR and therefore we generated a slightly larger deletion, Df(1)ED13157, a 288-kb deficiency removing the 18 genes between CG32529 and CG9576, which was also male viable. We identified three other nonvital regions of the fly genome during the course of our screening, all of which were molecularly confirmed by PCR. In the 64B region, a deletion encompassing 7 genes between CG11357 and CG32246 [Df(3L)ED4342/Df(3L)ED210 trans-heterozygous combination] is viable. The trans-heterozygous combination Df(3L)ED4502/Df(3L)ED4543 is a viable deletion including the 9 genes between Meics and CG9040 in the 70C region. Finally, Df(3L)ED4079 is a homozygous viable deletion of the 4 genes Lsp1γ, CG13405, CG12483, and Pk61C in the 61A region.
Analysis of deletion construction:
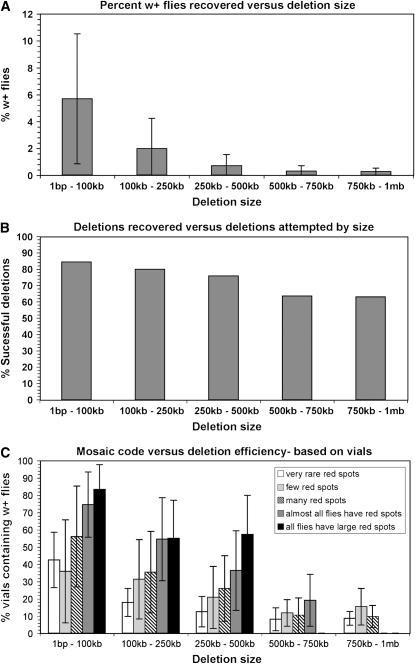
The frequency of deletion recovery was monitored in two different ways: either by absolute number of w+ flies recovered or by the number of vials that produced a w+ fly. The second method was preferred since it removes any bias resulting from germline clusters. Results of the deletion recovery screens analyzed by deletion size are summarized in Figure 5. Although the frequency of deletion recovery has a large standard deviation, there is a clearly observable trend between the frequency of recovery and the size of the deletion attempted (Figure 5A). These data also indicate that, although recovery of larger deletions requires screening larger numbers of progeny, there is only a slight difference in the overall success rate (Figure 5B); our observations here are similar to those reported by Golic and Golic (1996). A difference in somatic variegation after the “flip-in” round of heat shock was also noted such that crosses producing RS3r/RS5r trans-heterozygotes with a greater frequency of eye-color mosaicism tended to yield deletions more frequently in the following generation (Figure 5C). Deletion recovery frequencies were similar on all chromosome arms (data not shown).
Figure 5.—
Statistics of deletion recovery. (A) Deletion recovery frequency depends on the size of the deletion attempted. The percentage of deletion progeny recovered (Y axis) for deletions in a given size range (X axis), the bars represent standard deviations. (B) Absolute deletion recovery is affected by size, but not to a great extent. The frequency of success in generating deletions in the given size range is given, irrespective of the number of progeny that needed to be screened. (C) The extent of somatic mosaicism in heat-shocked flies carrying the RSr chromosomes in trans is a good indicator of successful deletion recovery. The frequency of deletion recovery (y-axis) for each of the indicated deletion size ranges is presented with respect to the subjective scoring of eye color mosaicism. In general parents with little mosaicism are less successful at producing deletion progeny.
Coverage of the DrosDel deletion kit:
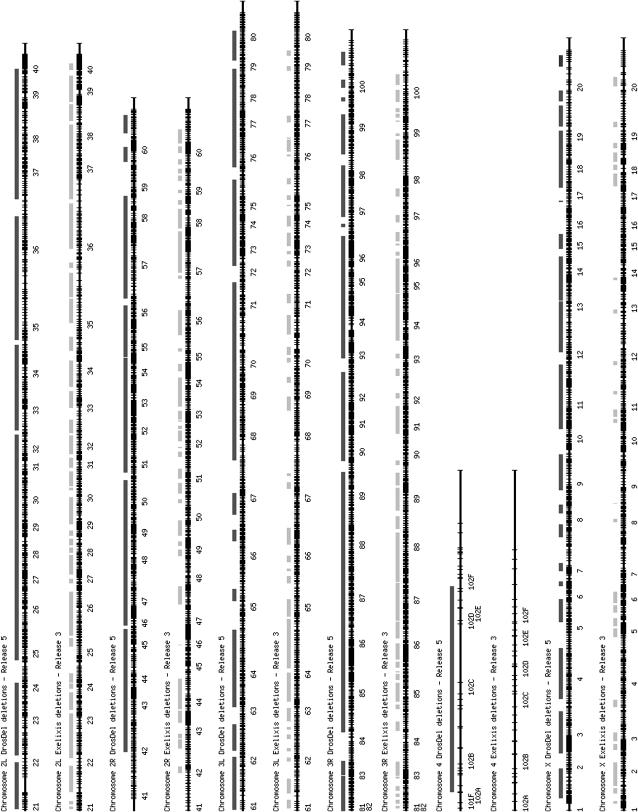
Current genome coverage of the DrosDel kit is summarized in Tables 1 and 2 and illustrated in Figure 6. A total of 665 deletions cover ∼77% of the Release 5.1 euchromatic genome sequence. Each deletion uncovers an average of 44 genes or 368 kb. The DrosDel deletions are, on average, 2.6 times larger than those in the collection produced by Parks et al. (2004), which has an average deletion size of 140 kb and a coverage of ∼56% of the genome. The different philosophies behind the design of the two collections offer complementary tools for groups investigating particular mutations or processes, since the DrosDel collection can be used for a “low-resolution” genome scan and the Exelixis collection for honing in, at higher resolution, on specific areas of interest highlighted in a DrosDel collection screen.
Figure 6.—
Genome coverage of DrosDel and Exelixis deletions. For each chromosome arm, the coverage of DrosDel deletions that have been made is mapped to the Release 5.1 genome sequence. The coverage of extant Exelixis deletions mapped to the Release 3 sequence is given below. In many cases, gaps are complementary. The extent of coverage on the autosomes is similar for both collections; however, DrosDel has considerably better representation on the X and also covers approximately half of chromosome 4.
At this time, a direct comparison between the DrosDel and Exelixis deletion collections with respect to the Release 5.1 genome sequence cannot be made since the sequence of some of the element insertion sites used to construct the Exelixis deletion set are not available. In Figure 6, we plot the DrosDel coverage on Release 5.1 compared with the coverage of Exelixis deletions still available from the stock center on Release 3 (of the 519 deletions originally reported in Parks et al. 2004, 452 remain alive and are in the Bloomington collection). While the exact breakpoints of the deletions plotted in Figure 6 will be different between Release 5.1 and 3 of the genome sequence, Figure 6 nicely illustrates an overall picture in which the two collections complement each other very well, with gaps in the DrosDel kit often being filled by the Exelixis collection and vice versa.
Duplications:
To increase the coverage of the DrosDel collection, and in particular to recover deletions in regions harboring haplo-insufficient loci, we set out to generate a series of duplication stocks. In addition to covering haplo-insufficient loci, duplications have a more general utility in dosage-sensitive screens. We describe here the general methods that we have adopted for duplication generation using the DrosDel collection. The start points for producing stable FRT-based duplications are inversions, which are generated by recombination between an RS3 and an RS5 element carried in cis and in the same relative orientation. To generate chromosomes with two RS elements, we selected a recombinant carrying both an RS3 and an RS5 by eye color, reduced the elements via the activity of FLP recombinase to generate a w chromosome, and carried out a second round of FLP-mediated recombination to generate a w+ inversion. An alternative method that we used was to make y w FLP; RS3r/RS5r females, cross these with y w FLP/y+Y;SM6/Sco males, and heat-shock the developing progeny. Male offspring showing mosaic eye colors had inherited a recombinant chromosome and these were used, after a further round of FLP treatment, to establish w; SM6/In stocks. Four inversion configurations are possible, depending upon the relative orientations of the starting RS elements (Figure 2A). Recombination in females trans-heterozygous for two inversions, having one similar breakpoint and one unique breakpoint, generates progeny with aneuploid chromosomes. For example, if a type 1 and type 2 inversion (w+) are combined (Figure 2B), the exceptional w progeny carry a duplication of region F, the region between the unique inversion breakpoints. The reciprocal event is a deletion, which can often be recognized phenotypically as a darker-eyed fly due to the two copies of w+ carried on this chromosome.
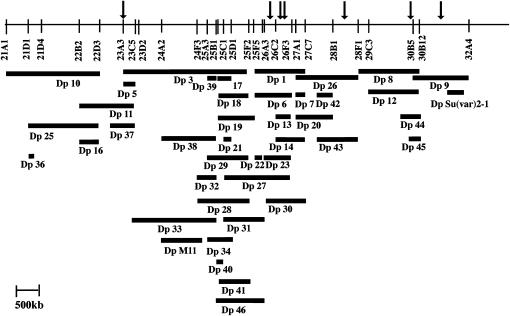
To illustrate the general utility of the DrosDel collection for carrying out this sophisticated chromosomal engineering, we focused on the distal half of chromosome arm 2L. We generated 48 inversion chromosomes (Table 3), designated In(2L)EINn (where n is a unique numerical identifier). In these particular examples, we have generated paracentric inversions; however, pericentric inversions are also easily generated (Golic and Golic 1996). We used these new inversions to generate a series of 41 duplication chromosomes [designated Dp(2;2)EDPn] covering the entirety of the chromosomal region from 21B1 to 32A4, ∼10% of the euchromatic genome (Figure 7 and Table 4). The duplications ranged in size from 20 kb [Dp(2;2)EDP36] to 2.91 Mb [Dp(2;2)EDP3], with high recovery rates (3–20% of progeny) that are apparently dependent upon the size of the duplications and the distance between the inversion breakpoints. To increase the utility of the duplication set, for example, for balancing haplo-lethal deletions, we selected pairs of inversions with different proximal breakpoints as well as the different distal breakpoints used to define the duplication. Thus all the duplication chromosomes carry a 334-kb deletion of the 37B1–C5 region and are homozygous lethal. Table 5 shows how the new duplications can be used to rescue otherwise inviable DrosDel deletions. For example, Dp(2;2)EDP5 allows the recovery and maintenance of six DrosDel deletions, which are all phenotypically Minute and lethal in combination with our preferred balancer, SM6a. In combination with Dp(2;2)EDP5, these deletions are healthy and not Minute. Similarly, Dp(2;2)EDP26 and Dp(2;2)EDP9 rescue Minutes at 28D and 31A, respectively. From similar crosses between inversions we also generated 17 new deletion chromosomes that we were able to maintain easily as stocks, 3 of which fill gaps in the standard deletion kit (Table 6). Taken together, this focused study illustrates how powerful FRT-based recombination can be for manipulating chromosomes with a high degree of accuracy. Again, we emphasize that all of the duplications are carried out in the same genetic background as the DrosDel deletions.
Figure 7.—
Duplication coverage for the distal half of 2L. A cytological map of the region of chromosome 2L from 21A1 to 32A4 with the location of the 41 duplications described in Table 4. Above the map, the locations of the lethal/haplo-insufficient regions rescued by the covering duplications described in Table 5 are indicated. Bar, 500 kb of genomic DNA.
TABLE 4.
2L Duplications
| Duplication | Inversion 1 | Inversion 2 | Duplicated region | Approximate size (Mb) |
|---|---|---|---|---|
| Dp(2;2)EDP10 | In(2L)EIN2 | In(2L)EIN31 | 21B1–22D3 | 1.13 |
| Dp(2;2)EDP36 | In(2L)EIN17 | In(2L)EIN30 | 21E2–21E2 | 0.02 |
| Dp(2;2)EDP25 | In(2L)EIN17 | In(2L)EIN31 | 21E2–22D3 | 1.60 |
| Dp(2;2)EDP16 | In(2L)EIN20 | In(2L)EIN31 | 22B2–22D3 | 0.46 |
| Dp(2;2)EDP11 | In(2L)EIN20 | In(2L)EIN23 | 22B2–23C4 | 1.28 |
| Dp(2;2)EDP37 | In(2L)EIN11 | In(2L)EIN23 | 22E1–23C4 | 0.72 |
| Dp(2;2)EDP5 | In(2L)EIN3 | In(2L)EIN23 | 23A3–23C4 | 0.26 |
| Dp(2;2)EDP3 | In(2L)EIN3 | In(2L)EIN35 | 23A3–25F2 | 2.91 |
| Dp(2;2)EDP33 | In(2L)EIN10 | In(2L)EIN22 | 23C5–25B1 | 1.83 |
| Dp(2;2)EDP38 | In(2L)EIN13 | In(2L)EIN22 | 24A2–25B1 | 1.26 |
| Dp(2;2)EDP32 | In(2L)EIN6 | In(2L)EIN22 | 24F3–25B1 | 0.44 |
| Dp(2;2)EDP28 | In(2L)EIN6 | In(2L)EIN35 | 24F3–25F2 | 1.20 |
| Dp(2;2)EDP39 | In(2L)EIN1 | In(2L)EIN22 | 25A3–25B1 | 0.19 |
| Dp(2;2)EDP34 | In(2L)EIN1 | In(2L)EIN33 | 25A3–25D1 | 0.54 |
| Dp(2;2)EDP29 | In(2L)EIN1 | In(2L)EIN35 | 25A3–25F2 | 0.96 |
| Dp(2;2)EDP40 | In(2L)EIN4 | In(2L)EIN27 | 25B1–25C1 | 0.14 |
| Dp(2;2)EDP17 | In(2L)EIN4 | In(2L)EIN33 | 25B1–25D1 | 0.34 |
| Dp(2;2)EDP18 | In(2L)EIN4 | In(2L)EIN35 | 25B1–25F2 | 0.77 |
| Dp(2;2)EDP46 | In(2L)EIN4 | In(2L)EIN28 | 25B1–26A3 | 1.06 |
| Dp(2;2)EDP19 | In(2L)EIN4 | In(2L)EIN29 | 25B1–26F3 | 1.76 |
| Dp(2;2)EDP21 | In(2L)EIN12 | In(2L)EIN33 | 25C3–25D1 | 0.18 |
| Dp(2;2)EDP31 | In(2L)EIN12 | In(2L)EIN28 | 25C3–26A3 | 0.89 |
| Dp(2;2)EDP27 | In(2L)EIN12 | In(2L)EIN29 | 25C3–26F3 | 1.60 |
| Dp(2;2)EDP22 | In(2L)EIN21 | In(2L)EIN28 | 25F5–26A3 | 0.15 |
| Dp(2;2)EDP6 | In(2L)EIN21 | In(2L)EIN29 | 25F5–26F3 | 0.85 |
| Dp(2;2)EDP1 | In(2L)EIN21 | In(2L)EIN26 | 25F5–27C7 | 1.16 |
| Dp(2;2)EDP23 | In(2L)EIN7 | In(2L)EIN29 | 26B1–26F3 | 0.67 |
| Dp(2;2)EDP30 | In(2L)EIN7 | In(2L)EIN26 | 26B1–27C7 | 0.98 |
| Dp(2;2)EDP41 | In(2L)EIN19 | In(2L)EIN29 | 26B2–26F3 | 0.65 |
| Dp(2;2)EDP13 | In(2L)EIN8 | In(2L)EIN29 | 26C1–26F3 | 0.38 |
| Dp(2;2)EDP14 | In(2L)EIN8 | In(2L)EIN26 | 26C1–27C7 | 0.69 |
| Dp(2;2)EDP7 | In(2L)EIN16 | In(2L)EIN26 | 27A1–27C7 | 0.25 |
| Dp(2;2)EDP20 | In(2L)EIN16 | In(2L)EIN34 | 27A1–28B1 | 0.87 |
| Dp(2;2)EDP26 | In(2L)EIN16 | In(2L)EIN32 | 27A1–28F1 | 1.50 |
| Dp(2;2)EDP42 | In(2L)EIN9 | In(2L)EIN34 | 27D3–28B1 | 0.57 |
| Dp(2;2)EDP43 | In(2L)EIN9 | In(2L)EIN32 | 27D3–28F1 | 1.20 |
| Dp(2;2)EDP8 | In(2L)EIN14 | In(2L)EIN25 | 28F1–30B12 | 1.38 |
| Dp(2;2)EDP12 | In(2L)EIN18 | In(2L)EIN25 | 29C3–30B12 | 1.17 |
| Dp(2;2)EDP44 | In(2L)EIN15 | In(2L)EIN25 | 30A4–30B12 | 0.38 |
| Dp(2;2)EDP45 | In(2L)EIN5 | In(2L)EIN25 | 30B3–30B12 | 0.14 |
| Dp(2;2)EDP9 | In(2L)EIN5 | In(2L)EIN24 | 30B3–32A5 | 1.30 |
Duplications were generated by recombination between the two inversions listed. The inversions are as described in Table 3. The approximate size of each duplication in megabases along with the predicted cytology is given. See text for details.
TABLE 5.
Rescuing haplo-insufficiency
| Duplication | Deficiency | Deletion cytology | Deletion size | Deletion phenotype |
|---|---|---|---|---|
| Dp(2;2)EDP5 | Df(2L)ED165 | 22F4–23B81 | 379,732 | Lethal over SM6a |
| Dp(2;2)EDP5 | Df(2L)ED183 | 23A3–23C21 | 156,582 | Lethal over SM6a |
| Dp(2;2)EDP5 | Df(2L)ED184 | 23A3–23C21 | 154,547 | Lethal over SM6a |
| Dp(2;2)EDP5 | Df(2L)ED195 | 23A3–23C41 | 265,279 | Lethal over SM6 |
| Dp(2;2)EDP5 | Df(2L)ED196 | 23A3–23C41 | 263,244 | Lethal over SM6a |
| Dp(2;2)EDP5 | Df(2L)ED167 | 22F4–23B81 | 380,892 | Lethal over SM6a |
| Dp(2;2)EDP3 | Df(2L)ED209 | 23A3–23C51 | 302,629 | Lethal over SM6a |
| Dp(2;2)EDP6 | Df(2L)ED389 | 25F5–26D7 | 663,854 | Weak over SM6a |
| Dp(2;2)EDP1 | Df(2L)ED378 | 26B2–26D7 | 465,644 | Weak over SM6a |
| Dp(2;2)EDP1 | Df(2L)ED6461 | 26C1–26F3 | 379,912 | Haplo-semilethal |
| Dp(2;2)EDP26 | Df(2L)ED522 | 28D3–28E12 | 94,779 | Minute |
| Dp(2;2)EDP9 | Df(2L)ED678 | 29F5–30B12 | 623,585 | Weak over SM6a |
| Dp(2;2)EDP9 | Df(2L)ED716 | 30E1–31B13 | 333,105 | Haplo-lethal |
Chromosome 2L duplications that can rescue otherwise inviable DrosDel deletions. Duplication descriptions are from Table 4. The relevant DrosDel deletion along with its size and predicted cytology are given. The phenotype of the heterozygous deletion is described in the phenotype column. Three of these regions are known to harbor Minute loci: 23B1, 28D2, and 31A3 (S. Marygold and K. Cook, personal communication).
TABLE 6.
Deletions from inversions
| Inversion 1 | Inversion 2 | Deficiency | Cytology | Size (Mb) |
|---|---|---|---|---|
| In(2L)EIN17 | In(2L)EIN31 | In(2L)EIN17L EIN31R | 21E2–22D3 | 1.61 |
| In(2L)EIN13 | In(2L)EIN22 | In(2L)EIN13L EIN22R | 24A2–25B1 | 1.26 |
| In(2L)EIN7 | In(2L)EIN29 | In(2L)EIN17L EIN29R | 26B1–26F3 | 0.67 |
New deletions were generated by recombination between inversions that cover gaps in the regular DrosDel deletion coverage. Inversions are described in Table 3.
Tip deletions:
To provide as complete a deletion kit as possible, we attempted to construct deletions covering the telomeric regions of each of the four major autosomal arms. These deletions were isolated by designing translocations in which terminal deletions were capped with the tip from another chromosome. The deletions were designed by selecting an RS5 “tip donor” element located very close to the tip of the X chromosome and corresponding RS3 “tip recipient” elements situated ∼100 kb from the tips of the autosomes. The tip donor element is in the minus orientation. The complementary tip-recipient elements are in the plus orientation on left arms or the minus orientation on right arms. These chromosomal aberrations are equivalent to the separable components of reciprocal translocations, i.e., translocation segregants (Ts). For example, where the tip of the X has been used to cap a terminal deletion of 2L, the resulting aberration could be described as Ts(1Lt;2Rt) because it carries the landmark telomeres from 1L and 2R. Four such tip deletions were isolated (Table 7), and in all cases the tip donor was the RS5 insertion 5-HA-1994, located in the minus orientation near the tip of the X (R5 sequence coordinate 19,199). Note that there are no known or predicted genes in the 19-kb duplicated region of the X chromosome. We tested the tip deletions by genetic complementation where possible: Df(2L)ED50001 failed to complement Df(2L)net-PMF or lethal alleles of l(2)gl, Df(3L)ED50003 failed to complement lethal alleles of krz, and Df(2R)ED50004 uncovered Kr. Thus, these deletions are confirmed genetically. Df(3L)ED50002 is homozygous viable and none of the five genes that it uncovers has a known visible or lethal phenotype. In all four cases, PCR confirmation of these deletions failed at the X-linked end only, suggesting a problem with the PCR conditions or the custom primer used for the 5-HA-1994 insertion, which could not be overcome.
TABLE 7.
Tip deletions
| Deletion | Recipient | Arm | Coordinate | Orientation | Size (bp) |
|---|---|---|---|---|---|
| Df(2L)ED50001 | CB-0264-3 | 2L | 72671 | Plus | 72671 |
| Df(2R)ED50004 | CB-0143-3 | 2R | 21113351 | Minus | 208059 |
| Df(3L)ED50002 | CB-5511-3 | 3L | 128631 | Plus | 128631 |
| Df(3R)ED50003 | CB-5616-3 | 3R | 27811479 | Minus | 875125 |
For construction of chromosome tip deletions, see text for details. The recipient RS element and its Release 5.1 scaffold location are given. The orientation of the recipient element and size of the terminal deletion are also indicated.
An FRT-derived deletion kit:
As we describe above, in addition to our DrosDel collection, the PiggyBac elements made by Exelixis (Thibault et al. 2004) have also been used to generate deletions by FRT-mediated recombination. While the collections are based on different transposable elements, they nevertheless contain very similar FRT sites. Therefore, in principle, it should be possible to combine elements from each collection to increase genome coverage and facilitate the generation of highly specific single-gene deletions. To facilitate such approaches, we calculated all possible deletions <500 kb that can be made by combining elements from the two collections and named these FDDs. To achieve this, we used the sequence data from the Exelixis collection of insertions from Harvard Medical School (http://drosophila.med.harvard.edu/) to remap these insertions with respect to the Release 5.1 genome sequence. Combining both collections, we find that over half a million (534,209) FDDs can theoretically be constructed (Table 8) and that >73,000 of these can be easily tracked through a change in eye color. The remaining 460,625 deletions can be detected by specific PCR assay. In total, these combined deletions cover >97% of the euchromatic Drosophila genome, although clearly there will still be some regions of the genome where deficiencies cannot be recovered due to haplo-insufficiency. However, in most cases, we have shown that deletions encompassing haplo-insufficient loci can readily be recovered by generating specific duplications. We therefore conclude that by combining both collections it will be possible to generate virtually complete genome coverage of precisely mapped deficiencies in defined genetic backgrounds. Drosophila therefore is the first model organism in which complete genetic dissection of a genome can be accomplished with the help of overlapping deletions, duplications, and other chromosomal rearrangements, all precisely defined at the DNA sequence level. This provides a powerful set of tools for comprehensive functional genomics with a complex eukaryotic genome.
TABLE 8.
FDD deletions
| Chromosome | Coverage (bp) | Chromosome length (bp) | % coverage | Deficiency: eye color | Deficiency: no color |
|---|---|---|---|---|---|
| 2L | 22,339,638 | 22,407,834 | 99.70 | 12,635 | 82,537 |
| 2R | 20,352,238 | 20,766,785 | 98.00 | 15,389 | 97,121 |
| 3L | 23,263,549 | 23,771,897 | 97.86 | 14,077 | 88,543 |
| 3R | 27,888,207 | 27,905,053 | 99.94 | 15,852 | 100,147 |
| 4 | 1,217,178 | 1,281,640 | 94.97 | 209 | 2,374 |
| X | 22,072,705 | 22,224,390 | 99.32 | 15,422 | 89,903 |
| Total | 117,133,515 | 118,357,599 | 98.97 | 73,584 | 460,625 |
This table is a summary of the FRT-derived deletions possible when the DrosDel and Exelixis collections of FRT-bearing inserts are combined. The total possible coverage for each chromosome arm is indicated as are the number of deletions that can be identified phenotypically (“eye color”) and those that require molecular identification (“no color”).
Using the FDD approach, we were interested in determining how many single-gene deletions could be constructed and found that a total of 614 complete single-gene deletions are possible; 30% of these can be easily tracked via an eye-color screen. In addition, a further 1704 partial-gene deletions, which would be expected to generate null alleles, can also be generated and 37% of these can be tracked by eye color. Taken together, we suggest that >15% of the predicted Drosophila gene complement could be disrupted with the FRT-based deletion approach. Of the 2318 gene disruptions that we predict, 14% have no known associated alleles. A database and a deletion search engine for FDDs are available at http://www.drosdel.org.uk/fdd/del_hunter.php.
DISCUSSION
Several years ago Golic and Golic (1996) demonstrated how recombination between FRT sites in the Drosophila genome could be used to precisely engineer chromosomes. In this article, we report the use of a collection of Drosophila stocks carrying FRT-containing P-elements to generate a large set of new chromosomal deletions. In addition, we show how the collection can be used to generate other chromosomal aberrations for manipulating the Drosophila genome. All of the starting elements are carried in an identical genetic background and are precisely mapped with respect to the genome. As a consequence, the breakpoints of all the chromosomal rearrangements that we have generated are accurately defined and the precise gene content of deleted or duplicated regions is known. This combination of genetic homogeneity and molecular precision is highly advantageous for genome-scale genetic screens and genomics studies (e.g., microarray experiments; Whitehead and Crawford 2006). Both are techniques where sensitivity to genetic background can result in many false positives or negatives; thus, eliminating background effects makes such screens less noisy. Similarly, when using deletions or duplications to carry out genetic or molecular dosage-sensitive screens, identification of contributing genes is expedited by knowing the gene content of aneuploid stocks. By making this collection available to the research community, we provide a set of tools that increase the already highly sophisticated way in which the fly genome can be manipulated and provide a technical route much easier to implement than more traditional chromosome engineering methods (e.g., Gubb 1998).
Demonstrating the utility of the collection, we generated a set of 642 deletions, covering 77% of the euchromatic genome and, as shown with chromosome arm 2L (Figure 3), the collection is capable of producing high-resolution tiles of overlapping deletions. In general, the FRT-based method appears to be robust when utilized at a whole-genome scale, a conclusion also reached by Park et al. (2004) when they developed a similar collection. The major barrier to generating full-genome coverage that we encountered was the issue of haplo-insufficiency or poor viability when the deletions are combined with common balancer chromosomes. In practice, these limitations prevent submission of our entire collection to the stock repositories, since healthy stocks are a prerequisite for high-volume fly maintenance. At present, we have made available a core collection of 209 validated and healthy stocks that cover >60% of the genome. Of course, as we demonstrate, recovery of a particular deletion is a relatively straightforward procedure and we are aware of 17 research groups that have utilized the DrosDel collection in published studies, using either our deletions or the tools and resources that we provide to generate custom deletions. To overcome this limitation, we demonstrate how stable covering duplications that rescue haplo-insufficiency can be easily generated from the DrosDel kit by FRT-mediated recombination. We have also demonstrated that the DrosDel kit can be used to identify previously unknown haplo-insufficient loci and to locate previously known loci on the scaffold. Our approach of using covering duplications complements the targeted hybrid element insertion and FRT-based methods being used by BDSC to generate deletions closely flanking haplo-insufficient loci (Parks et al. 2004).
The dominant male sterility of Df(3R)ED5647, Df(3R)ED5653, and Df(3R)ED10555 and the complete fertility of Df(3R)ED5664 has led us to the discovery and probable identification of a haplo-insufficient locus on chromosome 3, which we have named Ms(3)88C. These deletions restrict the male sterile region to 88C9;88D1 (R3 scaffold 10451431–10523038). The candidate genes in this region are His4r, Cad88C, CG7886, CG7832, CG3505, Rad17, CG3509, and Neu3 and we suggest that CG7866, which has ESTs expressed in the Drosophila testis (Andrews et al. 2000), is the most likely candidate.
Duplications allow the recovery of deletions of Minute loci, which usually correspond to haplo-insufficient ribosomal protein (Rp) genes (Lambertsson 1998; S. Marygold and K. Cook, personal communication). In the distal half of 2L, we constructed duplications to rescue otherwise inviable deletions in seven regions, including three regions known to harbor Minute loci: 23B, 28D, and 31A.
The DrosDel deletions were also used by S. Marygold and K. Cook (personal communication) to map Minute loci and hence aid in identification of the Rp genes that correspond to these Minutes. In mapping M(1)8F, it was noted that neither Df(1)ED7289 nor Df(1)ED7294 show a Minute phenotype. This delimits the number of candidate genes to just two, one of which is an Rp gene, RpL37a. Df(3R)ED6231 has a Minute phenotype and is the only deletion to uncover M(3)96CF. The deletion removes RpL27 and no other Rp gene, strongly suggesting the correspondence of this Minute locus with the Rp.
These results show how the DrosDel collection can be effectively employed to allow genetic analysis of even “difficult” regions of the genome. We continue to generate duplication chromosomes; we have currently generated almost complete coverage for 2L and approximately two-thirds of 2R and have started work on chromosome 3 (G. Reuter, unpublished data). The hope is that complete genome coverage will be obtained, facilitating both region-specific genetic analysis and genomewide dosage-sensitive screens.
Finally, the possibility of combining elements from the DrosDel and Exelixis collections offers the prospect of substantially increasing the genome coverage of small, precisely defined deletions. Together, these resources will facilitate very rapid and straightforward genetic analysis of defined regions of the Drosophila genome.
Acknowledgments
We thank Kevin Cook and Steven Marygold for sharing data and for their helpful discussions. Our thanks also to the Bloomington Drosophila Stock Center, the Szeged Drosophila Stock Centre, and the Exelixis Stock Collection at Harvard Medical School for providing fly strains. This work was supported by a European Union Framework Program 5 grant (contract no. QLRI-CT-2000-00915), by a Medical Research Council program grant (G8225539) to M.A. and S.R., and by Deutsche Forschungsgemeinschaft funding for G.R.
Note added in proof: Four hundred eighty five of the deletions reported in this article may now be obtained from the Szeged Stock Centre by ordering them from the DrosDel website. In addition, 312 deletions are available directly from the Bloomington Stock Center.
References
- Andrews, J., G. G. Bouffard, C. Cheadle, J. Lu, K. G. Becker et al., 2000. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 10: 2030–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ashburner, M., K. G. Golic and R. Scott Hawley, 2005. Drosophila: A Laboratory Handbook, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Golic, K. G., and M. M. Golic, 1996. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 144: 1693–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubb, D., 1998. Chromosome mechanics: the genetic manipulation of aneuploid stocks, pp. 109–130 in Drosophila: A Practical Approach, Ed. 2, edited by D. B. Roberts. Oxford University Press, New York.
- Kaiser, K., and S. F. Goodwin, 1990. “Site-selected” transposon mutagenesis of Drosophila. Proc. Natl. Acad. Sci. USA 87: 1686–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertsson, A., 1998. The minute genes in Drosophila and their molecular functions. Adv. Genet. 38: 69–134. [DOI] [PubMed] [Google Scholar]
- Lefevre, G., 1976. A photographic representation and interpretation of the polytene chromosomes of Drosophila melanogaster salivary glands, pp. 31–66 in The Genetics and Biology of Drosophila, Vol. 1a, edited by M. Ashburner and E. Novitski. Academic Press, London.
- Muller, H. J., 1930. Types of visible variations induced by X-rays in Drosophila. J. Genet. 22: 299–334. [Google Scholar]
- Parks, A. L., K. R. Cook, M. Belvin, N. A. Dompe, R. Fawcett et al., 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Peterson, H. M., and J. R. Laughnan, 1963. Intrachromosomal exchange at the Bar locus in Drosophila. Proc. Natl. Acad. Sci. USA 50: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S, and H. Skaletsky, 2000. Primer 3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Ryder, E., F. Blows, M. Ashburner, R. Bautista-Llacer, D. Coulson et al., 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., 1925. The effects of unequal crossing over at the Bar locus in Drosophila. Genetics 10: 117–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287. [DOI] [PubMed] [Google Scholar]
- Tsubota, S. I., 1991. Unequal crossing over within the B duplication of Drosophila melanogaster: a molecular analysis. Genet. Res. 57: 105–111. [DOI] [PubMed] [Google Scholar]
- Whitehead, A., and D. L. Crawford, 2006. Variation within and among species in gene expression: raw material for evolution. Mol. Ecol. 15: 1197–1211. [DOI] [PubMed] [Google Scholar]