Abstract
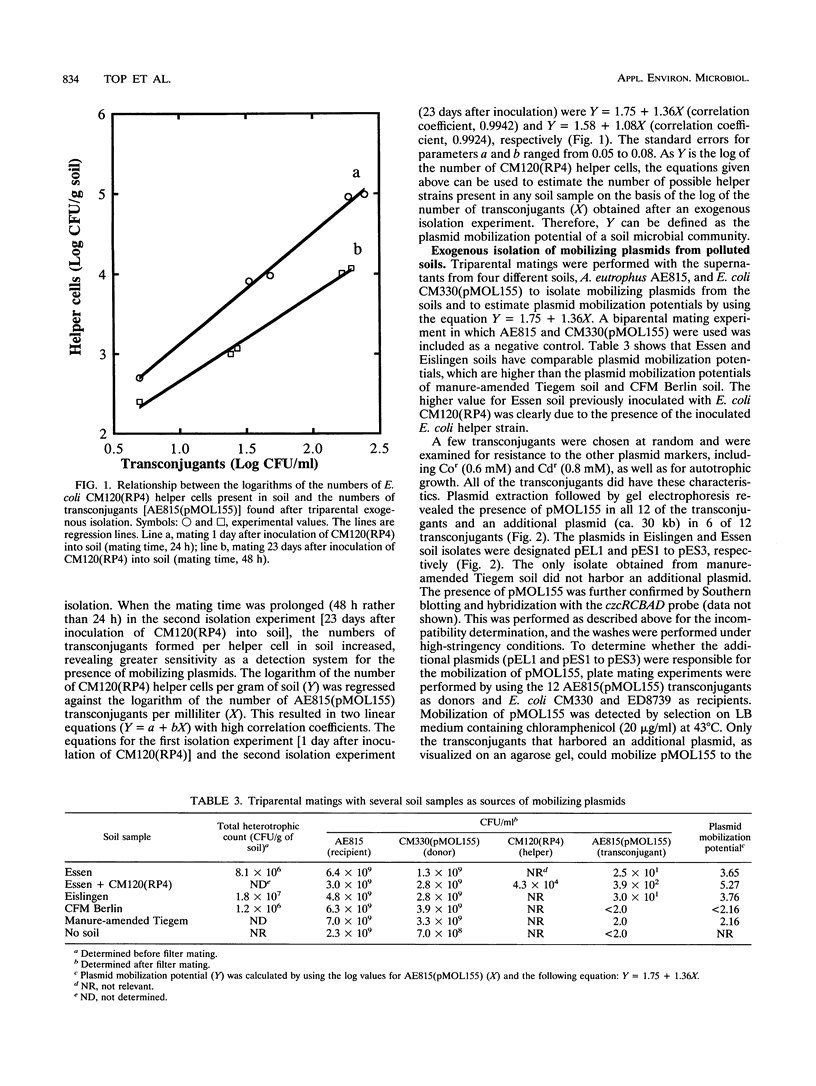
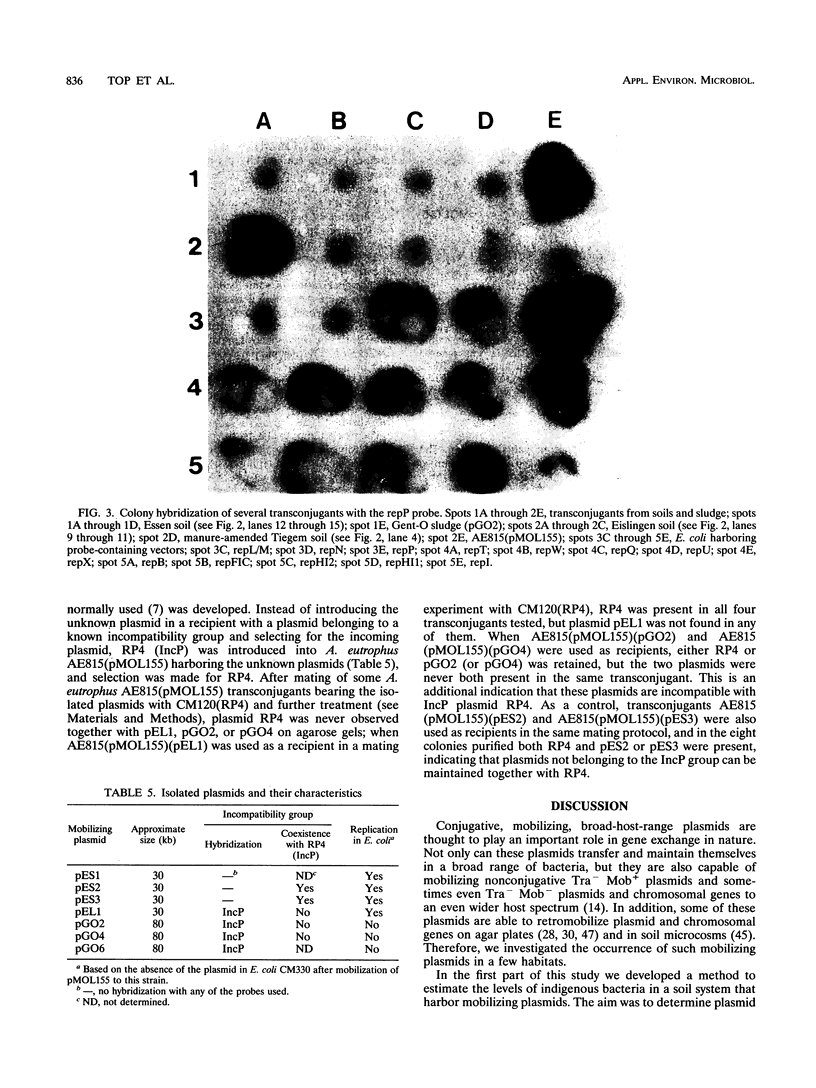
Exogenous plasmid isolation was used to assess the presence of mobilizing plasmids in several soils and activated sludges. Triparental matings were performed with Escherichia coli (a member of the γ subgroup of the Proteobacteria) as the donor of an IncQ plasmid (pMOL155, containing the heavy metal resistance genes czc: Cor, Znr, and Cdr), Alcaligenes eutrophus (a member of the β subgroup of the Proteobacteria) as the recipient, and indigenous microorganisms from soil and sludge samples as helper strains. We developed an assay to assess the plasmid mobilization potential of a soil ecosystem on the basis of the number of transconjugants obtained after exogenous isolations. After inoculation into soil of several concentrations of a helper strain (E. coli CM120 harboring IncP [IncP1] mobilizing plasmid RP4), the log numbers of transconjugants obtained from exogenous isolations with different soil samples were a linear function of the log numbers of helper strain CM120(RP4) present in the soils. Four soils were analyzed for the presence of mobilizing elements, and mobilizing plasmids were isolated from two of these soils. Several sludge samples from different wastewater treatment plants yielded much higher numbers of transconjugants than the soil samples, indicating that higher numbers of mobilizing strains were present. The mobilizing plasmids isolated from Gent-O sludge and one plasmid isolated from Eislingen soil hybridized to the repP probe, whereas the plasmids isolated from Essen soil did not hybridize to a large number of rep probes (repFIC, repHI1, repH12, repL/M, repN, repP, repT, repU, repW, repX). This indicates that in Essen soil, broad-host-range mobilizing plasmids belonging to other incompatibility groups may be present.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bale M. J., Fry J. C., Day M. J. Plasmid transfer between strains of Pseudomonas aeruginosa on membrane filters attached to river stones. J Gen Microbiol. 1987 Nov;133(11):3099–3107. doi: 10.1099/00221287-133-11-3099. [DOI] [PubMed] [Google Scholar]
- Bale M. J., Fry J. C., Day M. J. Transfer and occurrence of large mercury resistance plasmids in river epilithon. Appl Environ Microbiol. 1988 Apr;54(4):972–978. doi: 10.1128/aem.54.4.972-978.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier M., Bex F., Bergquist P. L., Maas W. K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988 Sep;52(3):375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley E. P., Davies J. Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J Bacteriol. 1991 Nov;173(21):6705–6708. doi: 10.1128/jb.173.21.6705-6708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hada H. S., Sizemore R. K. Incidence of Plasmids in Marine Vibrio spp. Isolated from an Oil Field in the Northwestern Gulf of Mexico. Appl Environ Microbiol. 1981 Jan;41(1):199–202. doi: 10.1128/aem.41.1.199-202.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Lehrbach P. R., Timmis K. N. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J Bacteriol. 1984 Oct;160(1):251–255. doi: 10.1128/jb.160.1.251-255.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K. E., Weightman A. J., Fry J. C. Isolation and screening of plasmids from the epilithon which mobilize recombinant plasmid pD10. Appl Environ Microbiol. 1992 Apr;58(4):1292–1300. doi: 10.1128/aem.58.4.1292-1300.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune P., Mergeay M., Van Gijsegem F., Faelen M., Gerits J., Toussaint A. Chromosome transfer and R-prime plasmid formation mediated by plasmid pULB113 (RP4::mini-Mu) in Alcaligenes eutrophus CH34 and Pseudomonas fluorescens 6.2. J Bacteriol. 1983 Sep;155(3):1015–1026. doi: 10.1128/jb.155.3.1015-1026.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini P., Fertels S., Nave D., Gealt M. A. Mobilization of plasmid pHSV106 from Escherichia coli HB101 in a laboratory-scale waste treatment facility. Appl Environ Microbiol. 1987 Apr;53(4):665–671. doi: 10.1128/aem.53.4.665-671.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure N. C., Weightman A. J., Fry J. C. Survival of Pseudomonas putida UWC1 containing cloned catabolic genes in a model activated-sludge unit. Appl Environ Microbiol. 1989 Oct;55(10):2627–2634. doi: 10.1128/aem.55.10.2627-2634.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson P., Gealt M. A. Isolation of indigenous wastewater bacterial strains capable of mobilizing plasmid pBR325. Appl Environ Microbiol. 1986 May;51(5):904–909. doi: 10.1128/aem.51.5.904-909.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M., Lejeune P., Sadouk A., Gerits J., Fabry L. Shuttle transfer (or retrotransfer) of chromosomal markers mediated by plasmid pULB113. Mol Gen Genet. 1987 Aug;209(1):61–70. doi: 10.1007/BF00329837. [DOI] [PubMed] [Google Scholar]
- Mergeay M., Nies D., Schlegel H. G., Gerits J., Charles P., Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985 Apr;162(1):328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Rafii F., Crawford D. L. Transfer of conjugative plasmids and mobilization of a nonconjugative plasmid between Streptomyces strains on agar and in soil. Appl Environ Microbiol. 1988 Jun;54(6):1334–1340. doi: 10.1128/aem.54.6.1334-1340.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre P., Miller R. V. Bacterial mobile genetic elements: importance in assessing the environmental fate of genetically engineered sequences. Plasmid. 1991 Nov;26(3):151–171. doi: 10.1016/0147-619x(91)90040-4. [DOI] [PubMed] [Google Scholar]
- Smit E., Venne D., van Elsas J. D. Mobilization of a Recombinant IncQ Plasmid between Bacteria on Agar and in Soil via Cotransfer or Retrotransfer. Appl Environ Microbiol. 1993 Jul;59(7):2257–2263. doi: 10.1128/aem.59.7.2257-2263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springael D., Kreps S., Mergeay M. Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J Bacteriol. 1993 Mar;175(6):1674–1681. doi: 10.1128/jb.175.6.1674-1681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Smith C. A. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- Top E., Mergeay M., Springael D., Verstraete W. Gene escape model: transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990 Aug;56(8):2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top E., Vanrolleghem P., Mergeay M., Verstraete W. Determination of the mechanism of retrotransfer by mechanistic mathematical modeling. J Bacteriol. 1992 Sep;174(18):5953–5960. doi: 10.1128/jb.174.18.5953-5960.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]