Abstract
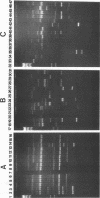
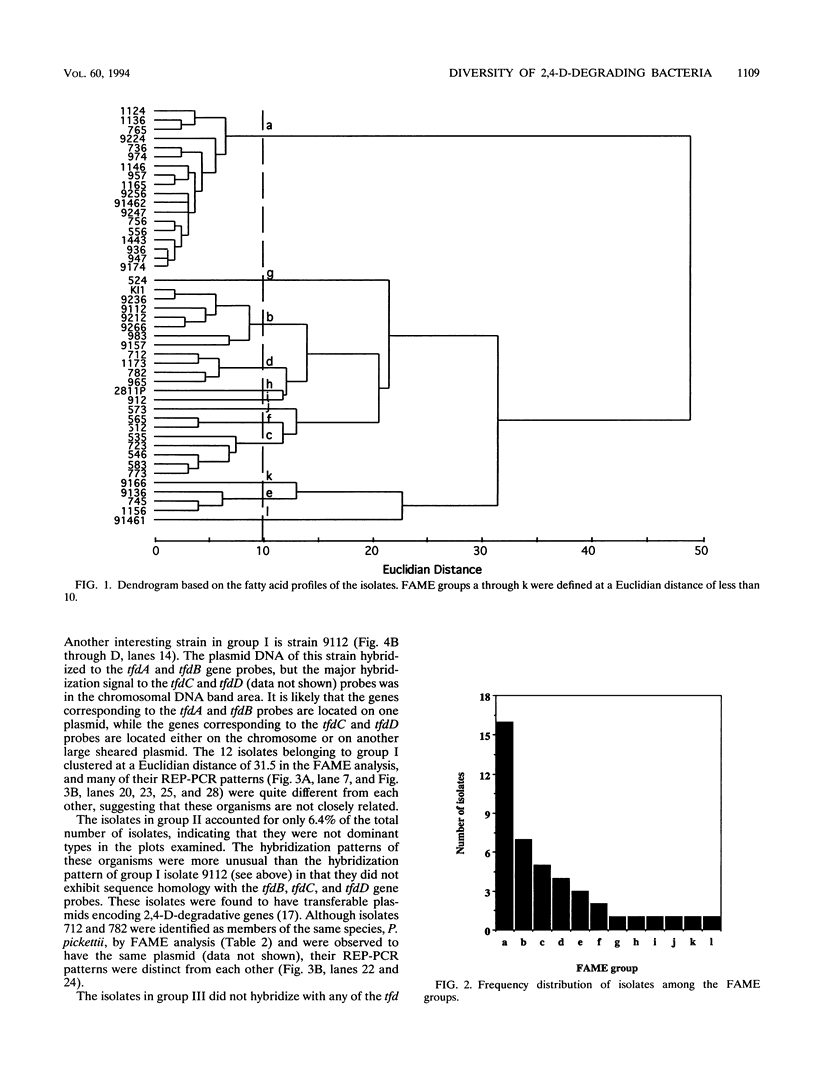
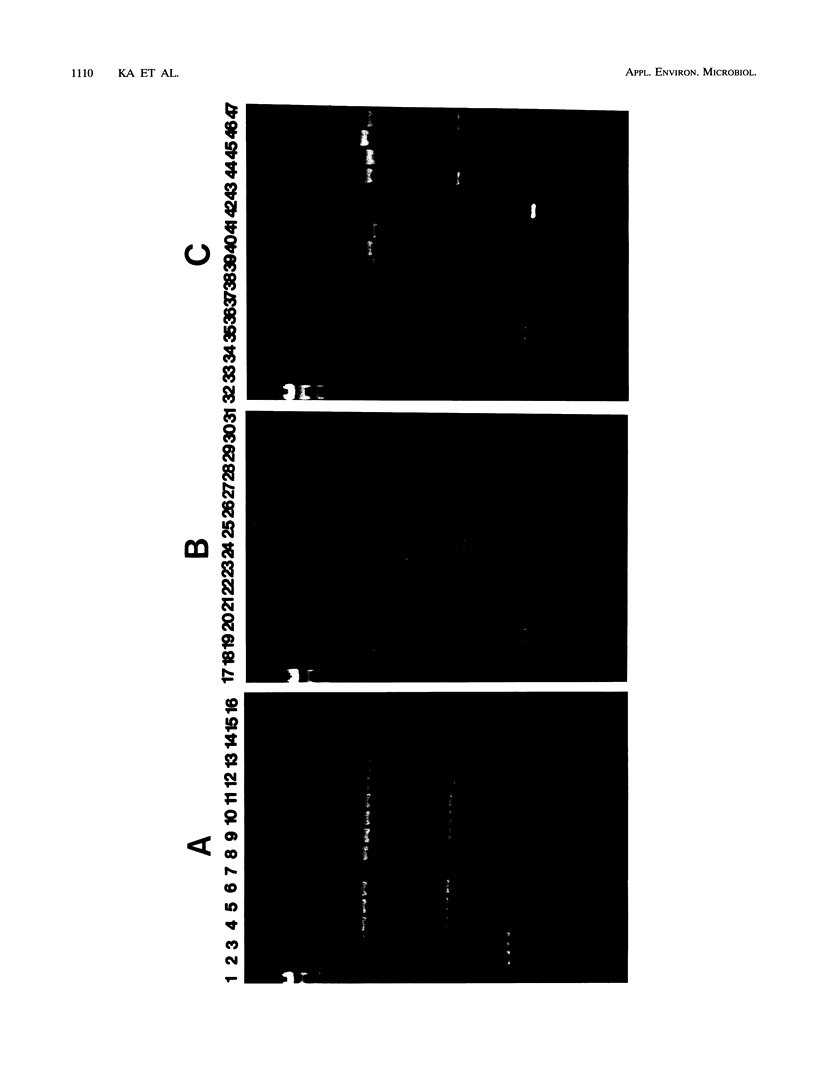
Forty-seven numerically dominant 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria were isolated at different times from 1989 through 1992 from eight agricultural plots (3.6 by 9.1 m) which were either not treated with 2,4-D or treated with 2,4-D at three different concentrations. Isolates were obtained from the most dilute positive most-probable-number tubes inoculated with soil samples from the different plots on seven sampling dates over the 3-year period. The isolates were compared by using fatty acid methyl ester (FAME) profiles, chromosomal patterns obtained by PCR amplification of repetitive extragenic palindromic (REP) sequences, and hybridization patterns obtained with probes for the tfd genes of plasmid pJP4 and a probe (Spa probe) that detects a distinctly different 2,4-D-degrading isolate, Sphingomonas paucimobilis (formerly Pseudomonas paucimobilis). A total of 57% of the isolates were identified to the species level by the FAME analysis, and these isolates were strains of Sphingomonas, Pseudomonas, or Alcaligenes species. Hybridization analysis revealed four groups. Group I strains, which exhibited sequence homology with tfdA, -B, -C, and -D genes, were rather diverse, as determined by both the FAME analysis and the REP-PCR analysis. Group II, which exhibited homology only with the tfdA gene, was a small group and was probably a subset of group I. All group I and II strains had plasmids. Hybridization analysis revealed that the tfd genes were located on plasmids in 75% of these strains and on the chromosome or a large plasmid in the other 25% of the strains. One strain exhibited tfdA and -B hybridization associated with a plasmid band, while tfdC and -D hybridized with the chromosomal band area. The group III strains exhibited no detectable homology to tfd genes but hybridized to the Spa probe. The members of this group were tightly clustered as determined by both the FAME analysis and the REP-PCR analysis, were distinctly different from group I strains as determined by the FAME analysis, and had very few plasmids; this group contained more of the 47 isolates than any other group. The group III strains were identified as S. paucimobilis. The group IV strains, which hybridized to neither the tft prove nor the Spa probe, were as diverse as the group I strains as determined by the FAME and REP-PCR analyses. Most of group IV strains could not be identified by the FAME analysis.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy P. S., Schulke J. W., Frazier L. M., Seidler R. J. Characterization of aquatic bacteria and cloning of genes specifying partial degradation of 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol. 1985 May;49(5):1237–1245. doi: 10.1128/aem.49.5.1237-1245.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELL G. R. Some morphological and biochemical characteristics of a soil bacterium which decomposes 2, 4-dichlorophenoxyacetic acid. Can J Microbiol. 1957 Oct;3(6):821–840. doi: 10.1139/m57-092. [DOI] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981 Feb;145(2):681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Weightman A. J., Knackmuss H. J., Timmis K. N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J Bacteriol. 1985 Jan;161(1):85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans W. C., Smith B. S., Fernley H. N., Davies J. I. Bacterial metabolism of 2,4-dichlorophenoxyacetate. Biochem J. 1971 May;122(4):543–551. doi: 10.1042/bj1220543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher P. R., Appleton J., Pemberton J. M. Isolation and characterization of the pesticide-degrading plasmid pJP1 from Alcaligenes paradoxus. J Bacteriol. 1978 Sep;135(3):798–804. doi: 10.1128/jb.135.3.798-804.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben W. E., Schroeter B. M., Calabrese V. G., Olsen R. H., Kukor J. K., Biederbeck V. O., Smith A. E., Tiedje J. M. Gene probe analysis of soil microbial populations selected by amendment with 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol. 1992 Dec;58(12):3941–3948. doi: 10.1128/aem.58.12.3941-3948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holben William E., Jansson Janet K., Chelm Barry K., Tiedje James M. DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community. Appl Environ Microbiol. 1988 Mar;54(3):703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JENSEN H. L., PETERSEN H. I. Detoxication of hormone herbicides by soil bacteria. Nature. 1952 Jul 5;170(4314):39–40. doi: 10.1038/170039a0. [DOI] [PubMed] [Google Scholar]
- Ka J. O., Holben W. E., Tiedje J. M. Analysis of competition in soil among 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl Environ Microbiol. 1994 Apr;60(4):1121–1128. doi: 10.1128/aem.60.4.1121-1128.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka J. O., Holben W. E., Tiedje J. M. Use of gene probes to aid in recovery and identification of functionally dominant 2,4-dichlorophenoxyacetic acid-degrading populations in soil. Appl Environ Microbiol. 1994 Apr;60(4):1116–1120. doi: 10.1128/aem.60.4.1116-1120.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos M. A., Roberts R. N., Alexander M. Phenols as intermediates in the decomposition of phenoxyacetates by an Arthrobacter species. Can J Microbiol. 1967 Jun;13(6):679–690. doi: 10.1139/m67-090. [DOI] [PubMed] [Google Scholar]
- NEWMAN A. S., WALKER R. L. Microbial decomposition of 2, 4-dichlorophenoxyacetic acid. Appl Microbiol. 1956 Jul;4(4):201–206. doi: 10.1128/am.4.4.201-206.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazinski J., Cen Y. H., Rolfe B. G. General method for the identification of plasmid species in fast-growing soil microorganisms. Appl Environ Microbiol. 1985 Apr;49(4):1001–1003. doi: 10.1128/aem.49.4.1001-1003.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROGOFF M. H., REID J. J. Bacterial decomposition of 2, 4-dichlorophenoxyacetic acid. J Bacteriol. 1956 Mar;71(3):303–307. doi: 10.1128/jb.71.3.303-307.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Streber W. R., Timmis K. N., Zenk M. H. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987 Jul;169(7):2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi E., Yano I., Oyaizu H., Hashimoto Y., Ezaki T., Yamamoto H. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol Immunol. 1990;34(2):99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x. [DOI] [PubMed] [Google Scholar]
- de Bruijn F. J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992 Jul;58(7):2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]