Abstract
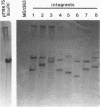
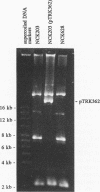
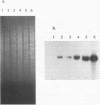
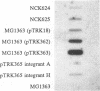
The abiA gene (formerly hsp) encodes an abortive phage infection mechanism which inhibits phage DNA replication. To analyze the effects of varying the abiA gene dosage on bacteriophage resistance in Lactococcus lactis, various genetic constructions were made. An IS946-based integration vector, pTRK75, was used to integrate a single copy of abiA into the chromosomes of two lactococcal strains, MG1363 and NCK203. In both strains, a single copy of abiA did not confer any significant phage resistance on the host except for one of the MG1363 integrants, NCK625, which exhibited a slightly higher level of resistance to phages sk1 and p2. Hybridization of the total cellular RNA from NCK625 to an abiA-specific probe indicated that the integration took place downstream of a promoter causing stronger expression of abiA in this integrant. Three abiA-containing plasmids of various copy numbers were introduced into both strains, and the recombinants were evaluated for resistance to phages c2, p2, sk1, and φ31. Plasmid pTRK18 has a copy number of approximately six (cn = 6) and caused a decreased plaque size for all phages evaluated. Integration of pTRK75 into a native plasmid of NCK203 generated pTRK362 (cn = 13), which caused a reduced efficiency of plaquing (EOP = 10-2) and reduced plaque size. A high-copy-number abiA plasmid (pTRK363), based on the pAMβ1 origin of replication, was also constructed (cn = 100). Plasmid pTRK363 caused a significant reduction in EOP (10-4 to 10-8) and plaque size for all phages tested, although in some cases, this plasmid caused the evolution of AbiA-resistant phage derivatives. Altering the gene dosage or expression level of abiA significantly affects the phage resistance levels.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alatossava T., Klaenhammer T. R. Molecular Characterization of Three Small Isometric-Headed Bacteriophages Which Vary in Their Sensitivity to the Lactococcal Phage Resistance Plasmid pTR2030. Appl Environ Microbiol. 1991 May;57(5):1346–1353. doi: 10.1128/aem.57.5.1346-1353.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C., Ehrlich S. D., Jannière L. Unidirectional theta replication of the structurally stable Enterococcus faecalis plasmid pAM beta 1. EMBO J. 1991 Aug;10(8):2171–2177. doi: 10.1002/j.1460-2075.1991.tb07752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J., Daly C., Fitzgerald G. F. Controlled Integration into the Lactococcus Chromosome of the pCI829-Encoded Abortive Infection Gene from Lactococcus lactis subsp. lactis UC811. Appl Environ Microbiol. 1992 Oct;58(10):3283–3291. doi: 10.1128/aem.58.10.3283-3291.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao M. L., Ferretti J. J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985 Jan;49(1):115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feirtag J. M., Petzel J. P., Pasalodos E., Baldwin K. A., McKay L. L. Thermosensitive plasmid replication, temperature-sensitive host growth, and chromosomal plasmid integration conferred by Lactococcus lactis subsp. cremoris lactose plasmids in Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1991 Feb;57(2):539–548. doi: 10.1128/aem.57.2.539-548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. L., Sanozky-Dawes R. B., Klaenhammer T. R. Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J Bacteriol. 1988 Aug;170(8):3435–3442. doi: 10.1128/jb.170.8.3435-3442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Massey I. J., Klaenhammer T. R. Rapid method to characterize lactococcal bacteriophage genomes. Appl Environ Microbiol. 1991 Jan;57(1):283–288. doi: 10.1128/aem.57.1.283-288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. Nucleotide sequence and distribution of the pTR2030 resistance determinant (hsp) which aborts bacteriophage infection in lactococci. Appl Environ Microbiol. 1990 Jul;56(7):2255–2258. doi: 10.1128/aem.56.7.2255-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. The bacteriophage resistance plasmid pTR2030 forms high-molecular-weight multimers in lactococci. Plasmid. 1991 Mar;25(2):105–112. doi: 10.1016/0147-619x(91)90021-n. [DOI] [PubMed] [Google Scholar]
- Hill C., Pierce K., Klaenhammer T. R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction modification (R+/M+) and abortive infection (Hsp+). Appl Environ Microbiol. 1989 Sep;55(9):2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Romero D. A., McKenney D. S., Finer K. R., Klaenhammer T. R. Localization, cloning, and expression of genetic determinants for bacteriophage resistance (Hsp) from the conjugative plasmid pTR2030. Appl Environ Microbiol. 1989 Jul;55(7):1684–1689. doi: 10.1128/aem.55.7.1684-1689.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horodniceanu T., Bouanchaud D. H., Bieth G., Chabbert Y. A. R plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother. 1976 Nov;10(5):795–801. doi: 10.1128/aac.10.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Conferred on Lactic Streptococci by the Conjugative Plasmid pTR2030: Effects on Small Isometric-, Large Isometric-, and Prolate-Headed Phages. Appl Environ Microbiol. 1986 Jun;51(6):1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- Le Chatelier E., Ehrlich S. D., Jannière L. Biochemical and genetic analysis of the unidirectional theta replication of the S. agalactiae plasmid pIP501. Plasmid. 1993 Jan;29(1):50–56. doi: 10.1006/plas.1993.1006. [DOI] [PubMed] [Google Scholar]
- Leenhouts K. J., Gietema J., Kok J., Venema G. Chromosomal stabilization of the proteinase genes in Lactococcus lactis. Appl Environ Microbiol. 1991 Sep;57(9):2568–2575. doi: 10.1128/aem.57.9.2568-2575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Stabilization of Lactose Metabolism in Streptococcus lactis C2. Appl Environ Microbiol. 1978 Aug;36(2):360–367. doi: 10.1128/aem.36.2.360-367.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'sullivan D. J., Hill C., Klaenhammer T. R. Effect of Increasing the Copy Number of Bacteriophage Origins of Replication, in trans, on Incoming-Phage Proliferation. Appl Environ Microbiol. 1993 Aug;59(8):2449–2456. doi: 10.1128/aem.59.8.2449-2456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya R. R., Kleeman E. G., Luchansky J. B., Klaenhammer T. R. Characterization of the temperate bacteriophage phi adh and plasmid transduction in Lactobacillus acidophilus ADH. Appl Environ Microbiol. 1989 Sep;55(9):2206–2213. doi: 10.1128/aem.55.9.2206-2213.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Klaenhammer T. R. Characterization of insertion sequence IS946, an Iso-ISS1 element, isolated from the conjugative lactococcal plasmid pTR2030. J Bacteriol. 1990 Aug;172(8):4151–4160. doi: 10.1128/jb.172.8.4151-4160.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Klaenhammer T. R. IS946-mediated integration of heterologous DNA into the genome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1992 Feb;58(2):699–702. doi: 10.1128/aem.58.2.699-702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Characterization of Phage-Sensitive Mutants from a Phage-Insensitive Strain of Streptococcus lactis: Evidence for a Plasmid Determinant that Prevents Phage Adsorption. Appl Environ Microbiol. 1983 Nov;46(5):1125–1133. doi: 10.1128/aem.46.5.1125-1133.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen J. M., van der Lelie D., Venema G. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl Environ Microbiol. 1987 Oct;53(10):2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]