Abstract
CTP synthetase is an essential enzyme that generates the CTP required for the synthesis of nucleic acids and membrane phospholipids. In this work, we examined the phosphorylation of the human CTPS1-encoded CTP synthetase 1 by protein kinase A. CTP synthetase 1 was expressed and purified from a Saccharomyces cerevisiae ura7Δ ura8Δ double mutant that lacks CTP synthetase activity. Using purified CTP synthetase 1 as a substrate, protein kinase A activity was time- and dose-dependent. The phosphorylation, which primarily occurred on a threonine residue, was accompanied by a 50% decrease in CTP synthetase 1 activity. The synthetic peptide LGKRRTLFQT that contains the protein kinase A motif for Thr455 was a substrate for protein kinase A. A Thr455 to Ala (T455A) mutation in CTP synthetase 1 was constructed by site-directed mutagenesis and was expressed and purified from the S. cerevisiae ura7Δ ura8Δ mutant. The T455A mutation caused a 78% decrease in protein kinase A phosphorylation, and the loss of the phosphothreonine residue and a major phosphopeptide that were present in the purified wild type enzyme phosphorylated by protein kinase A. The CTP synthetase 1 activity of the T455A mutant enzyme was 2-fold higher than the wild type enzyme. In addition, the T455A mutation caused a 44% decrease in the amount of human CTP synthetase 1 that was phosphorylated in S. cerevisiae cells, and this was accompanied by a 2.5-fold increase in the cellular concentration of CTP and a 1.5-fold increase in the choline-dependent synthesis of phosphatidylcholine.
CTP synthetase (EC 6.3.4.2, UTP: ammonia ligase (ADP-forming)) catalyzes the ATP-dependent transfer of the amide nitrogen from glutamine (i.e., glutaminase reaction) to the C-4 position of UTP to generate CTP (1, 2). GTP activates the glutaminase reaction by accelerating the formation of a covalent glutaminyl enzyme intermediate (2, 3). The enzyme displays positive cooperative kinetics with respect to UTP and ATP, and negative cooperative kinetics with respect to glutamine and GTP (2–10). The kinetic behavior with respect to UTP and ATP is attributed to the nucleotide-dependent tetramerization of the enzyme (2, 8, 11, 12). In fact, it is the tetrameric form of the enzyme that is active (2–10, 12).
CTP synthetase is an essential enzyme for life because it catalyzes the formation of the CTP required for the synthesis of nucleic acids and membrane phospholipids (13). In eukaryotic cells, CTP synthetase activity controls the balance of nucleotide pools (8, 14–19) and regulates the synthesis of membrane phospholipids (19–21). The importance of understanding the regulation of CTP synthetase is highlighted by the fact that an unregulated level of its activity is a phenotype common to leukemia cells (22–24) and rapidly growing tumors of liver (25), colon (26), and lung (27). Indeed, CTP synthetase is a target of antiproliferative drug development for cancer therapy (28–34), and a target for parasite- (35) and viral-based (36) diseases.
Two important modes of CTP synthetase regulation are feedback inhibition by CTP (2, 8–10, 19, 37) and covalent modification by phosphorylation (38–40). A defect in CTP feedback inhibition of CTP synthetase activity causes elevated intracellular levels of CTP (15, 19, 41), resistance to nucleotide analog drugs used in cancer chemotherapy (42–45), and increased rates of spontaneous mutations (16, 43, 45). The yeast URA7-encoded CTP synthetase is phosphorylated on serine residues by protein kinases A (39) and C (38, 46). These modifications facilitate the nucleotide-dependent tetramerization of CTP synthetase (12), attenuate the sensitivity of the enzyme to feedback inhibition by CTP (38, 39), and stimulate enzyme activity (38, 39, 46).
In humans, CTP synthetase is encoded by the CTPS1 (47) and CTPS2 (48) genes, which are located on chromosomes 1p34 and Xp22, respectively. The CTPS1- and CTPS2-encoded CTP synthetases have predicted molecular masses of 66.7 and 65.7 kDa, respectively, and show 74% identity in their amino acid sequences (47, 48). CTP synthetase 1 contains conserved synthetase and glutamine amide transfer domains that are involved in enzyme catalysis (49–51) (Fig. 1). Human CTP synthetase 1 shares a relatively high degree of amino acid sequence identity (~53%) with the yeast URA7- and URA8-encoded CTP synthetase enzymes (17, 18). In fact, either one of the human CTP synthetase enzymes is functional in yeast and complements the lethal phenotype of a ura7Δ ura8Δ double mutant lacking CTP synthetase activity (40). In addition, the expression of human CTP synthetase 1 in yeast has revealed that the enzyme is phosphorylated by protein kinase A (40). The specificity of this phosphorylation has been addressed through in vitro studies using Escherichia coli-expressed pure CTP synthetase 1 and mammalian protein kinase A (40).
FIGURE 1. Domain structure of human CTP synthetase 1 and putative protein kinase A phosphorylation sites.
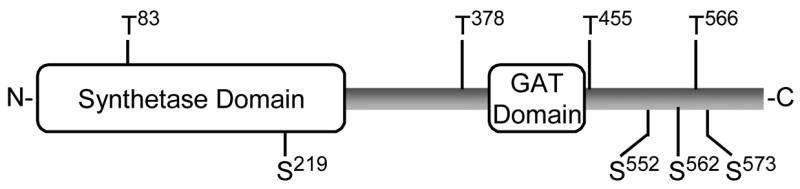
The diagram shows the positions of the synthetase domain (residues 1–272) and the glutamine amide transfer (GAT) domain (residues 394–405) in the CTP synthetase 1 protein sequence. The putative serine (S) and threonine (T) residues for protein kinase A phosphorylation are indicated.
In this work, we examined the protein kinase A phosphorylation of the human CTP synthetase 1 that was expressed and purified from yeast. Unlike the yeast CTP synthetase that is stimulated by protein kinase A phosphorylation (39), the human enzyme activity was inhibited by protein kinase A. We identified Thr455 as a major site of protein kinase A phosphorylation. Analysis of a T455A mutant CTP synthetase 1 indicated that phosphorylation at Thr455 resulted in the inhibition of activity in vitro and in vivo. In addition, data indicated that phosphorylation at Thr455 attenuates the choline-dependent synthesis of phosphatidylcholine when CTP synthetase 1 enzyme was expressed in S. cerevisiae. The identification of Thr455 as a major protein kinase A target site advances our understanding of human CTP synthetase 1 regulation by phosphorylation.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were reagent grade. Supplies for the growth of yeast and bacteria were purchased from Difco Laboratories. New England Biolabs was the source of restriction endonucleases, modifying enzymes, and recombinant Vent DNA polymerase. Kits for the purification of plasmid DNA and the extraction of DNA from agarose gels, and Ni2+-NTA agarose resin were purchased from Qiagen. DNA size ladder used for agarose gel electrophoresis was from Invitrogen. Oligonucleotides were synthesized at Genosys Biotechnologies, Inc. Peptides were synthesized and purified commercially by BioSynthesis, Inc. Yeast transformation kit was obtained from Clontech. The QuikChange site-directed mutagenesis kit was purchased from Stratagene. PerkinElmer Life Sciences was the source of radiochemicals. Aprotinin, benzamidine, bovine serum albumin, 5-fluoroorotic acid, leupeptin, lyticase, nucleotides, pepstatin, phenylmethylsulfonyl fluoride, standard phosphoamino acids, and L-1-tosylamido-2-phenylethyl chloromethyl ketone-trypsin were purchased from Sigma. Protein assay reagents, electrophoretic reagents, and protein standards for SDS-PAGE were purchased from Bio-Rad. Mouse monoclonal anti-His6 antibodies and alkaline phosphatase-conjugated goat anti-mouse antibodies were purchased from Cell Signaling Technology and Pierce, respectively. Protein kinase A catalytic subunit (bovine heart) was purchased from Promega. Hybond-P PVDF membrane and the enhanced chemifluorescence Western blotting detection kit were purchased from Amersham Biosciences. Silica Gel 60 thin-layer and cellulose thin-layer chromatography plates were purchased from EM Science. Scintillation counting supplies were from National Diagnostics.
Strains and Growth Conditions
S. cerevisiae cells were grown in synthetic complete (SC) medium containing 2% glucose at 30 °C as described previously (52, 53). For selection of cells bearing plasmids, appropriate amino acids were omitted from SC medium. Cells were also grown in YEPD medium (1% yeast extract, 2% peptone, 2% glucose). Plasmid maintenance and amplifications were performed in Escherichia coli strain DH5α. E. coli cells were grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.4) at 37 °C. Ampicillin (100 μg/ml) was added to the growth medium for bacteria carrying plasmids. The growth media were supplemented with either 2% (yeast) or 1.5% (E. coli) agar for growth on plates. Growth of cultures in liquid media was determined spectrophotometrically at A600 nm.
DNA Manipulations, Amplification of DNA by PCR, and DNA Sequencing
Standard methods were used to prepare genomic and plasmid DNA, to digest DNA with restriction enzymes, and to ligate DNA (54). Standard methods were also used for the transformation of yeast and E. coli (53, 55, 56). PCR reactions were optimized as described previously (57). DNA sequencing reactions were performed by the dideoxy method using Taq polymerase (53) and analyzed with an automated DNA sequencer.
Construction of Plasmids and Expression of CTPS1 Mutant Alleles in S. cerevisiae
The plasmids and oligonucleotide primers used in this work are listed in Tables 2 and 3, respectively. Mutant alleles of CTPS1 were constructed by PCR with the QuikChange site-directed mutagenesis kit using plasmid pDO105-hCTPS1 (40) as the template. This is a LEU2-based plasmid that directs the overexpression of full-length His6-tagged (C terminus) human CTP synthetase 1 in S. cerevisiae (40). The CTPS1T455A,S552A mutant allele was constructed with the primers for the S552A mutation using plasmid CTPS1T455A as the template. All mutations were confirmed by DNA sequencing.
TABLE 2.
Plasmids used in this work
| Plasmid | Relevant characteristics | Source or Ref |
|---|---|---|
| YEpLac181 | Multicopy E. coli/yeast shuttle vector with LEU2 | (83) |
| YEpLac195 | Multicopy E. coli/yeast shuttle vector with URA3 | (83) |
| pDO105 | Derivative of YEpLac181 with the ADH1 promoter and multiple cloning sites | (19) |
| pDO134 | URA7 derivative of YEpLac195 | (19) |
| pDO105-hCTPS1 | CTPS1 derivative of pDO105 | (40) |
| pDO105-hCTPS1(T83A) | CTPS1T83A derivative of pDO105-hCTPS1 | This study |
| pDO105-hCTPS1(S219A) | CTPS1S219A derivative of pDO105-hCTPS1 | This study |
| pDO105-hCTPS1(T378A) | CTPS1T378A derivative of pDO105-hCTPS1 | This study |
| pDO105-hCTPS1(T455A) | CTPS1T455A derivative of pDO105-hCTPS1 | This study |
| pDO105-hCTPS1(S552A) | CTPS1S552A derivative of pDO105-hCTPS1 | This study |
| pDO105-hCTPS1(S562A) | CTPS1S562A derivative of pDO105-hCTPS1 | This study |
| pDO105-hCTPS1(T566A) | CTPS1T566A derivative of pDO105-hCTPS1 | This study |
| pDO105-hCTPS1(S573A) | CTPS1S573A derivative of pDO105-hCTPS1 | This study |
| pDO105-hCTPS1(T455A,S552A) | CTPS1T455A,S552A derivative of pCTPS T455A | This study |
TABLE 3.
Oligonucleotides used in this work
| Oligo | Sequence |
|---|---|
| T83A Fa | 5′-CTTGACATCCGCCTCGCCAAGGACAATAATCTG-3′ |
| T83A Rb | 5′-CAGATTATTGTCCTTGGCGAGGCGGATGTCAAG-3′ |
| S219A F | 5′-GTTGTATGCAGGTGCGCAAATCCACTTGACAC-3′ |
| S219A R | 5′-GTGTCAAGTGGATTTGCGCACCTGCATACAAC-3′ |
| T378A F | 5′-GATTTGGTGTTCGAGGAGCAGAAGGAAAAATCC-3′ |
| T378A R | 5′-GGATTTTTCCTTCTGCTCCTCGAACACCAAATC-3′ |
| T455A F | 5′-CTGGGCAAGAGGAGAGCCCTGTTCCAGACCAAG-3′ |
| T455A R | 5′-CTTGGTCTGGAACAGGGCTCTCCTCTTGCCCAG-3′ |
| S552A F | 5′-CTGTGGGGCGGCTCGCACATTACCTCCAG-3′ |
| S552A R | 5′-CTGGAGGTAATGTGCGAGCCGCCCCACAG-3′ |
| S562A F | 5′-GAAAGGCTGCAGGCTCGCACCCAGGGACACCTATAG-3′ |
| S562A R | 5′-CTATAGGTGTCCCTGGGTGCGAGCCTGCAGCCTTTC-3′ |
| T566A F | 5′-CTCTCACCCAGGGACGCCTATAGTGACAGG-3′ |
| T566A R | 5′-CCTGTCACTATAGGCGTCCCTGGGTGAGAG-3′ |
| S573A F | 5′-GTGACAGGAGTGGAGCCAGCTCCCCTGACTC-3′ |
| S573A R | 5′-GAGTCAGGGGAGCTGGCTCCACTCCTGTCAC-3′ |
Forward orientation relative to sense strand
Reverse orientation relative to sense strand
Strain SDO195 was transformed to leucine prototrophy with plasmids containing the mutant alleles of the CTPS1 gene. Strain SDO195 is a ura7Δ ura8Δ double mutant bearing plasmid pDO134. This plasmid, which bears a wild type URA7 allele, rescues the lethal phenotype of the ura7Δ ura8Δ mutant (18), and at the same time, confers uracil prototrophy to strain SDO195 because it also bears the URA3 gene (19). The yeast transformants, which contained URA3- and LEU2-based plasmids bearing the URA7 and CTPS1 genes, respectively, were streaked onto SC-leucine plates containing 5-fluoroorotic acid to evict (58) the plasmid bearing the URA3 gene. 5-Fluoroorotic acid-resistant cells were further confirmed by their uracil auxotrophy. The 5-fluoroorotic acid-resistant cells, which are ura7Δ ura8Δ mutant cells expressing the wild type and mutant CTPS1 genes, showed growth that was indistinguishable from that of cells expressing the yeast URA7 gene.
Purification of Human CTP Synthetase 1 from S. cerevisiae
All steps were performed at 4 °C. Cells expressing His6-tagged wild type and T455A mutant human CTP synthetase 1 enzymes were used for enzyme purification. Freshly harvested cells from 1-liter cultures were disrupted with glass beads using a Biospec Products Bead-Beater in 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.3 M sucrose, 10 mM β-mercaptoethanol, 0.5 mM phenylmethanesulfonyl fluoride, 1 mM benzamidine, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 5 μg/ml pepstatin, 10 mM NaF, and 5 mM β-glycerophosphate. Glass beads and unbroken cells were removed by centrifugation at 1,500 × g for 5 min to obtain the cell extract. The cell extract was centrifuged at 100,000 × g for 1.5 h to obtain the cytosolic fraction. The cytosolic fraction was loaded onto a 1 ml Ni2+-NTA column. The column was washed with 20 mM Tris-HCl (pH 7.9), 0.5 M NaCl, 45 mM imidazole, 10% glycerol, and 7 mM β-mercaptoethanol to remove nonspecific proteins. The His6-tagged enzyme was eluted from the column with 20 mM Tris-HCl (pH 7.9), 0.5 M NaCl, 250 mM imidazole, 10% glycerol, and 7 mM β-mercaptoethanol. Fractions containing the enzyme were dialyzed against 20 mM Tris-HCl (pH 7.9), 10% glycerol, and 7 mM β-mercaptoethanol, and concentrated using an Amicon Ultra Centrifugal filter. For small-scale purification, wild type and mutant enzymes were purified directly from cell extracts using Ni2+-NTA resin. Cell extract (0.5 mg protein) was incubated with a 10% slurry (w/v) of Ni2+-NTA resin for 1 h at 4 °C. The Ni2+-NTA resin-bound enzyme was collected by centrifugation and used for phosphorylation experiments. Analysis by SDS-PAGE indicated that the enzyme preparations purified by both procedures were nearly homogeneous.
Enzyme Assays
CTP synthetase activity was determined by measuring the conversion of UTP to CTP (molar extinction coefficients of 182 and 1520 M−1 cm−1, respectively) by following the increase in absorbance at 291 nm on a recording spectrophotometer (2). The standard reaction mixture contained 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 10 mM β-mercaptoethanol, 2 mM L-glutamine, 0.1 mM GTP, 2 mM ATP, 2 mM UTP, and an appropriate dilution of enzyme protein in a total volume of 0.1 ml. Alternatively, the CTP synthetase reaction product CTP was determined by high performance liquid chromatography using the method of Mole et al. (59). Enzyme reactions were terminated by the addition of 0.1 ml of 0.26 M ammonium phosphate/acetonitrile (10:1, pH 5.55). The reaction mixtures were then filtered through Microcon-30 centrifuge filters. Samples (0.1 ml) were then subjected to analytical high performance liquid chromatography using a Partisil 10 SAX column (250 × 4.6 mm, inner diameter) with a SAX guard column (60). The column was equilibrated and eluted with 0.26 M ammonium phosphate/acetonitrile (10:1, pH 5.55) at a flow rate of 2 ml/min. The identity of CTP was determined by comparing elution profiles with that of standard CTP using an ultraviolet detector (A265 nm). The concentration of CTP was determined from a standard curve. Enzyme assays were performed in triplicate with an average standard deviation of ± 3%. All assays were linear with time and protein concentration. A unit of CTP synthetase activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of product/min.
SDS-PAGE and Immunoblot Analysis
SDS-PAGE (61) and immunoblotting (62) with PVDF membrane were carried out as described previously. Mouse monoclonal anti-His6 antibodies and alkaline phosphatase-conjugated goat anti-mouse antibodies were used at dilutions of 1:1000 and 1:5000, respectively. CTP synthetase 1 was detected on immunoblots using the enhanced chemifluorescence Western blotting detection kit as described by the manufacturer. The images from the immunoblots were acquired by FluorImaging. The relative densities of the images were analyzed using ImageQuant software, and the signals were in the linear range of detectability.
Phosphorylation of Human CTP Synthetase 1 and Synthetic Peptides with Protein Kinase A
Phosphorylation reactions were performed in a total volume of 25 μl at 30 °C. Purified CTP synthetase 1 (ca. 1 μg) was incubated for 10 min with 50 mM Tris-HCl (pH 8.0), 60 mM β-mercaptoethanol, 10 mM MgCl2, 50 μM [γ-32P]ATP (5 μCi/nmol), and the indicated concentrations of mammalian protein kinase A catalytic subunit. At the end of the phosphorylation reactions samples were treated with 4x Laemmli’s sample buffer (61), subjected to SDS-PAGE, and transferred to PVDF membranes. The phosphorylated enzyme was visualized by phosphorimaging, and the extent of phosphorylation was quantified with ImageQuant software. The data were normalized to amount of human CTP synthetase 1 protein on the membranes as determined by immunoblot analysis. Reactions using human CTP synthetase 1 synthetic peptides were performed in a total volume of 40 μl, and were terminated by spotting 20 μl onto P81 phosphocellulose paper. The papers were washed with 75 mM phosphoric acid and then subjected to scintillation counting. Phosphorylation reactions were performed in triplicate. A unit of protein kinase A activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of product/min.
In Vivo Labeling of Human CTP synthetase 1
The S. cerevisiae ura7Δ ura8Δ mutant bearing the wild type and mutant human CTPS1 alleles was used to examine the phosphorylation of the human CTP synthetase 1 enzyme. Cultures (50 ml) in SC medium were grown to the exponential phase of growth. Cells were harvested and resuspended in 5 ml of fresh medium containing 32Pi (0.25 mCi/ml) and incubated for 3 h. The 32P-labeled cells were harvested by centrifugation, suspended in 0.25 ml of spheroplast buffer (25 mM Tri-HCl (pH 7.4), 1 M sorbitol, 5 mM dithiothreitol, 10 mM NaF, 10 mM NaN3, and 30 units of lyticase), and incubated for 30 min at 30 °C. The spheroplasts were lysed by boiling for 3 min in 2% SDS. His6-tagged human CTP synthetase 1 enzymes were isolated from the lysates with Ni2+-NTA resin, subjected to SDS-PAGE, and transferred to PVDF membrane. The membrane was used for phosphorimaging analysis of 32P-labeled human CTP synthetase 1 enzymes. ImageQuant software was used to quantify the extent of enzyme phosphorylation. The data were normalized to amount of human CTP synthetase 1 protein on the membranes as determined by immunoblot analysis using anti-His6 antibodies.
Phosphoamino Acid Analysis and Phosphopeptide Mapping Analysis
For phosphoamino acid analysis, a portion of a PVDF membrane containing 32P-labeled human CTP synthetase 1 protein was subjected to acid hydrolysis with 6 N HCl (39). Hydrolysates were dried in vacuo and applied to cellulose thin-layer chromatography plates with standard phosphoamino acids (2.5 μg of phosphoserine, 2.5 μg of phosphothreonine, and 5 μg of phosphotyrosine) and separated by electrophoresis using formic acid/acetic acid/water (50:156:1794, v/v) in the first dimension (pH 1.9 buffer) and acetic acid/pyridine/water (100:10:1890, v/v) in the second dimension (pH 3.5 buffer) (63). Following electrophoresis, the cellulose thin-layer plates were dried and subjected to phosphorimaging analysis. Standard phosphoamino acids were visualized by spraying the plate with 0.25% ninhydrin in acetone.
For phosphopeptide mapping analysis, PVDF membrane slices containing 32P-labeled CTP synthetase 1 protein were subjected to digestion with L-1-tosylamido-2-phenylethyl chloromethyl ketone-trypsin and to two-dimensional peptide mapping analysis (64). Peptides were separated by electrophoresis (1% ammonium bicarbonate buffer at 1000V for 45 min) and by ascending chromatography (n-butyl alcohol/glacial acetic acid/pyridine/water, 10:3:12:15, for 8 h) on cellulose thin-layer chromatography plates. Dried plates were then subjected to phosphorimaging analysis.
Extraction and Analysis of CTP
Nucleotides were extracted from S. cerevisiae cells with 1 M formic acid/10% 1-butanol (v/v) as described previously (17). The nucleotide extract (0.1 ml) was analyzed for CTP by high performance liquid chromatography (60).
Analysis of Phospholipids
Exponential-phase yeast cells expressing the wild type and mutant human CTP synthetase 1 enzymes were incubated with 32Pi or with [methyl-14C]choline for 30 min to label phospholipids (65, 66). 32Pi is incorporated into phospholipids synthesized by both CDP-diacylglycerol-dependent and CDP-choline-dependent pathways, whereas [methyl-14C]choline is incorporated into phosphatidylcholine synthesized by only the CDP-choline-dependent pathway. Phospholipids were extracted (67) and analyzed by two-dimensional thin-layer chromatography on silica gel thin-layer chromatography plates (68). The solvent systems for dimensions one and two were chloroform/methanol/ammonium hydroxide/water (45:25:2:3, v/v) and chloroform/methanol/acetic acid/water (64:8:10:2, v/v), respectively. The radiolabeled phospholipids were visualized by phosphorimaging analysis. The positions of the labeled phospholipids on chromatography plates were compared with standard phospholipids after exposure to iodine vapor. The relative amounts of 32P-labeled phospholipids were quantified using ImageQuant software whereas the amount of 14C-labeled phosphatidylcholine was determined by liquid scintillation counting.
Analyses of Data
Statistical analyses were performed with SigmaPlot 7 software.
RESULTS AND DISCUSSION
Protein Kinase A Phosphorylation of S. cerevisiae-expressed Human CTP Synthetase 1, and the Effect of Protein Kinase A Phosphorylation on CTP Synthetase 1 Activity
Human CTP synthetase 1 was first identified as a phosphoprotein in a large-scale characterization of HeLa cell nuclear phosphoproteins (69). Covalent modification by phosphorylation is a major mechanism by which the activity of an enzyme is regulated (70, 71). Our phosphorylation studies on the human CTP synthetase 1 have been facilitated by the functional expression of the human CTPS1 gene in the S. cerevisiae ura7Δ ura8Δ double mutant that lacks CTP synthetase activity (40). In vivo labeling experiments using the yeast surrogate have confirmed that CTP synthetase 1 is phosphorylated, and that some of this phosphorylation is mediated by protein kinase A (40). Protein kinase A is a serine/threonine kinase, and the principal target of the second messenger cAMP (72). That CTP synthetase 1 is a substrate for mammalian protein kinase A has been established by in vitro studies using CTP synthetase 1 expressed and purified from E. coli (40).
The direct effect of protein kinase A phosphorylation on CTP synthetase 1 activity has been unclear because the E. coli-expressed enzyme has negligible activity, and phosphorylation does not restore activity (40). We have reasoned that the E. coli-expressed CTP synthetase 1 is inactive because the enzyme is not subject to all of the post-translational modifications required for maximum expression of activity (40). On the other hand, when human CTPS1 is expressed in S. cerevisiae ura7Δ ura8Δ cells, the activity of CTP synthetase 1 is comparable to that of the yeast URA7-encoded CTP synthetase enzyme (40). In this work, His6-tagged human CTP synthetase 1 was expressed and purified from S. cerevisiae. Analysis by SDS-PAGE indicated that the CTP synthetase 1 preparation used in this study was purified to apparent homogeneity (Fig. 2). The purified S. cerevisiae-expressed CTP synthetase 1 was phosphorylated by mammalian protein kinase A, and this phosphorylation was dependent on the time of the reaction and on the concentration of the kinase (Fig. 2). The effect of mammalian protein kinase A on purified CTP synthetase 1 activity is also shown in Fig. 2. Phosphorylation of CTP synthetase 1 resulted in a dose-dependent decrease in enzyme activity. At the concentration of 0.5 unit of protein kinase A, the activity of CTP synthetase 1 was reduced by 53%.
FIGURE 2. Phosphorylation of S. cerevisiae-expressed human CTP synthetase 1 by protein kinase A, and the effect of protein kinase A phosphorylation on CTP synthetase 1 activity.
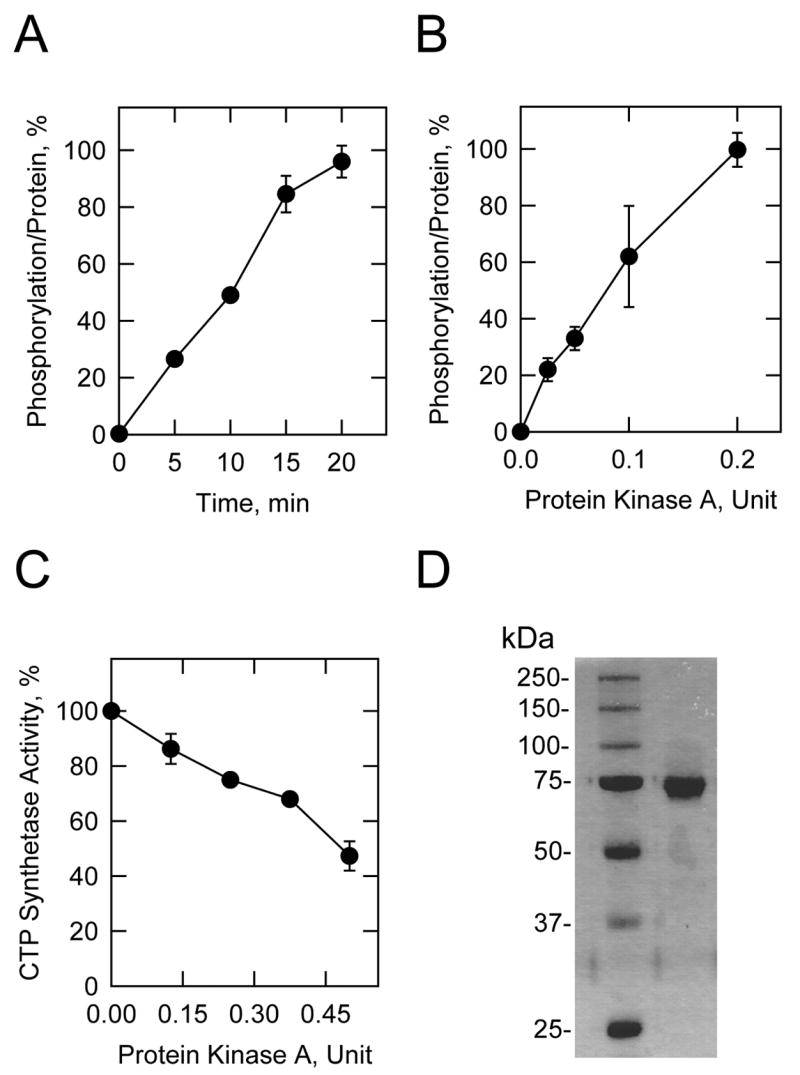
A, purified wild type human CTP synthetase 1 was incubated with mammalian protein kinase A (0.1 U/ml) and [γ-32P]ATP for the indicated time intervals. B, purified wild type human CTP synthetase 1 was incubated with [γ-32P]ATP and the indicated amounts of mammalian protein kinase A for 10 min. After incubations, samples were subjected to SDS-PAGE and transferred to PVDF membrane. The phosphorylated proteins were subjected to phosphorimaging analysis and the relative amounts of phosphate incorporated were quantified using ImageQuant software. The maximum extent of CTP synthetase 1 phosphorylation was set at 100%. The data were normalized to the amount of human CTP synthetase 1 protein as determined by immunoblot analysis using anti-His6 antibodies. The values reported were average of four separate experiments ± S.D. C, purified human CTP synthetase 1 was phosphorylated for 20 min with the indicated amounts of mammalian protein kinase A using unlabeled ATP. The control reaction contained unlabeled ATP and omitted protein kinase A. The product CTP was extracted from the reaction mixture and analyzed by high performance liquid chromatography. The CTP synthetase activity measured without protein kinase A phosphorylation was set at 100%. The values reported were the average of three separate experiments ± S.D. D, purified wild type CTP synthetase 1 was subjected to SDS-PAGE and stained with Coomassie blue. The positions of the protein molecular mass standards are indicated on the left side of the figure.
Human CTP synthetase 1 Synthetic Peptides Containing a Protein Kinase A Sequence Motif Are Substrates for Protein Kinase A
Identification of the protein kinase A phosphorylation site(s) in CTP synthetase 1 was examined to gain insight into the relevance of enzyme phosphorylation. Analysis of the human CTP synthetase 1 sequence with the NetPhosK 1.0 server (http://www.cbs.dtu.dk/services/NetPhosK/) (73) indicated that the enzyme has potential protein kinase A phosphorylation sites at four serine (Ser219, Ser552, Ser562, and Ser573) and four threonine (Thr83, Thr378, Thr455, and Thr566) residues (Fig. 1). Peptides containing each of the putative protein kinase A target sites were synthesized and tested for their ability to serve as substrates for mammalian protein kinase A. Of the eight peptides, only peptides T455 and S552 served as substrates; the protein kinase A activity using peptide T455 was 4-fold greater than the activity using peptide S552 (Table 4). The incorporation of the γ-phosphate of ATP into the T455 and S552 peptides by protein kinase A was dependent on the concentration of each peptide (Fig. 3). The peptides A455 and A552 were synthesized and tested as substrates for protein kinase A phosphorylation (Table 4). These peptides did not serve as substrates for protein kinase A, indicating that the threonine and serine residues contained within the protein kinase A motif of the peptides T455 and S552 were the target sites for protein kinase A phosphorylation.
TABLE 4.
Protein kinase A activity using human CTP synthetase 1 synthetic peptides
| Peptidea | Sequenceb | Protein kinase A activity |
|---|---|---|
| nmol/min/mg | ||
| T83 | LDIRLTKDNN | NAc |
| S219 | VVCRCSNPLD | 0.57 ± 0.1 |
| T378 | FGVRGTEGKI | 0.45 ± 0.2 |
| T455 | LGKRRTLFQT | 107.1 ± 20 |
| A455 | LGKRRALFQT | NA |
| S552 | SVGRLSHYLQ | 27 ± 1 |
| A552 | SVGRLAHYLQ | NA |
| S562 | QKGCRLSPRD | NA |
| T566 | PRDTYSDRSG | 0.11 ± 0.1 |
| S573 | YSDRSGSSSP | NA |
Protein kinase A activity measured with 0.5 mM peptide.
Putative phosphorylation site is underlined
No activity.
FIGURE 3. Phosphorylation of human CTP synthetase 1 synthetic peptides by protein kinase.
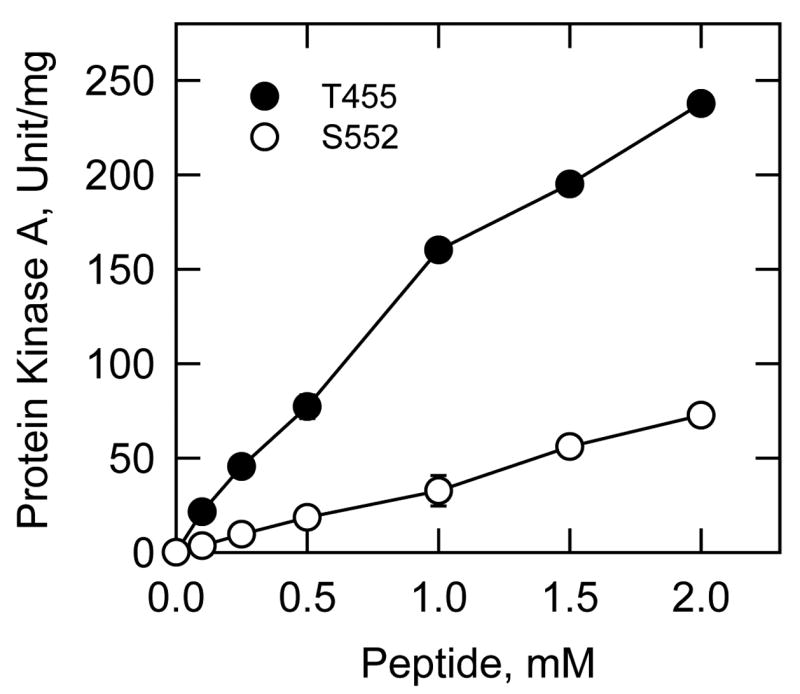
A. Mammalian protein kinase A activity was measured as a function of the concentration of the indicated synthetic peptides T455 and S552. The values reported were the average of two separate experiments ± S.D.
Effects of Protein Kinase A Phosphorylation Site Mutations on the Phosphorylation of Human CTP Synthetase 1 by Protein Kinase A
In complementary studies, four serine (Ser219, Ser552, Ser562, and Ser573) and four threonine (Thr83, Thr378, Thr455, and Thr566) residues within the human CTP synthetase 1 were mutagenized to further test the hypothesis that these sites may be targets for mammalian protein kinase A phosphorylation. CTPS1 alleles with serine-to-alanine and threonine-to-alanine mutations were constructed by site-directed mutagenesis, and expressed as His6-tagged proteins. The protein kinase A phosphorylation site mutant alleles were expressed from a multicopy plasmid in a ura7Δ ura8Δ mutant to obviate any effects due to the CTP synthetase activities encoded by the native yeast URA7 (17) and URA8 (18) genes. A multicopy plasmid was used for increased expression to facilitate purification of the mutant His6-tagged enzymes. Immunoblot analysis using anti-His6 antibodies showed that the mutations did not affect the expression levels of the human CTP synthetase 1 enzyme in yeast. In addition, the mutations in the CTP synthetase 1 did not affect the growth of ura7Δ ura8Δ cells.
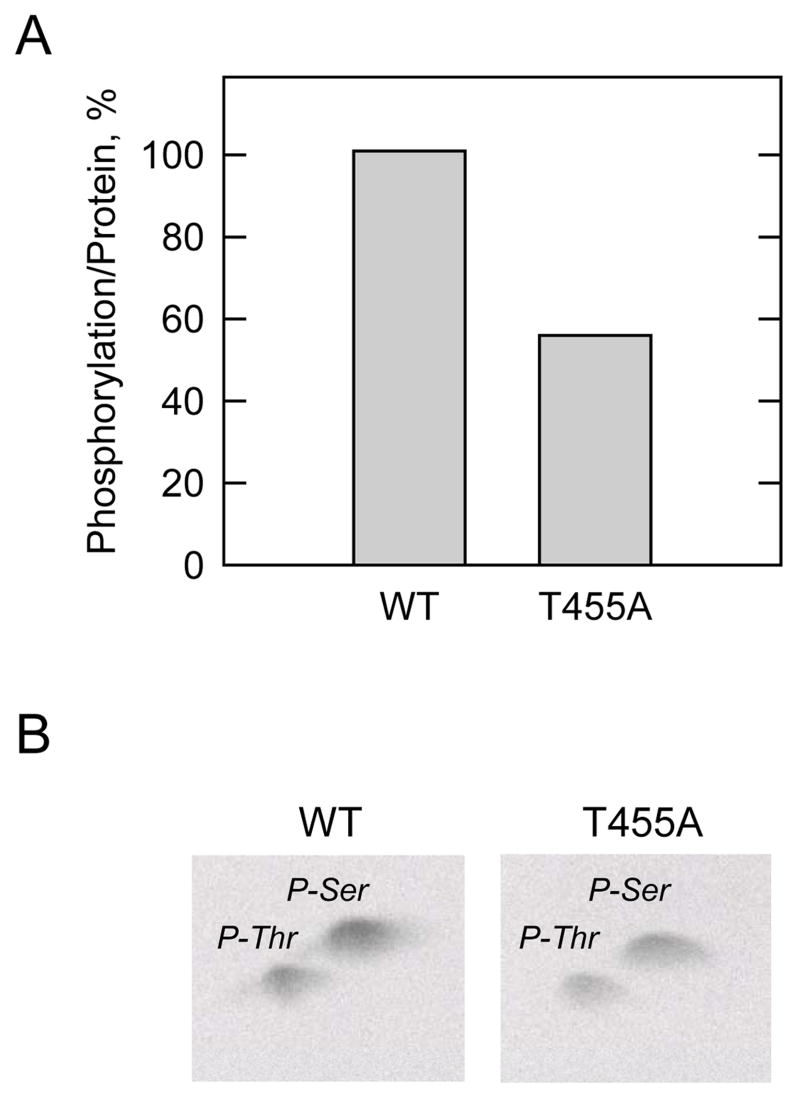
The mutant His6-tagged proteins were isolated from yeast cell extracts using Ni2+-NTA agarose resin, and the resin-bound proteins were then incubated with mammalian protein kinase A and [γ-32P]ATP. After the phosphorylation reactions, samples were subjected to SDS-PAGE, transferred to PVDF membrane, and then analyzed for the radioactive label incorporated into human CTP synthetase 1. The amounts of the mutant proteins on the membrane were confirmed by immunoblot analysis using anti-His6 antibodies. Of all the mutant proteins tested, only the T455A mutant showed a defect in protein kinase A phosphorylation. The extent of phosphorylation of the T455A was reduced by 78% when compared with the wild type enzyme (Fig. 4A). When human CTP synthetase 1 is expressed and purified from E. coli, protein kinase A primarily phosphorylates the enzyme on a serine residue(s) (40). However, when the enzyme was expressed and purified from S. cerevisiae, protein kinase A primarily phosphorylated CTP synthetase 1 on a threonine residue(s) (Fig. 4B). The reason for this difference is unclear, but may be related to posttranslational modification(s) of the protein that occurred in yeast but not in E. coli.
FIGURE 4. Effects of the T455A, S552A, and T455A,S552A mutations on the phosphorylation of human CTP synthetase 1 by protein kinase A, and phosphoamino acid analysis of phosphorylated proteins.
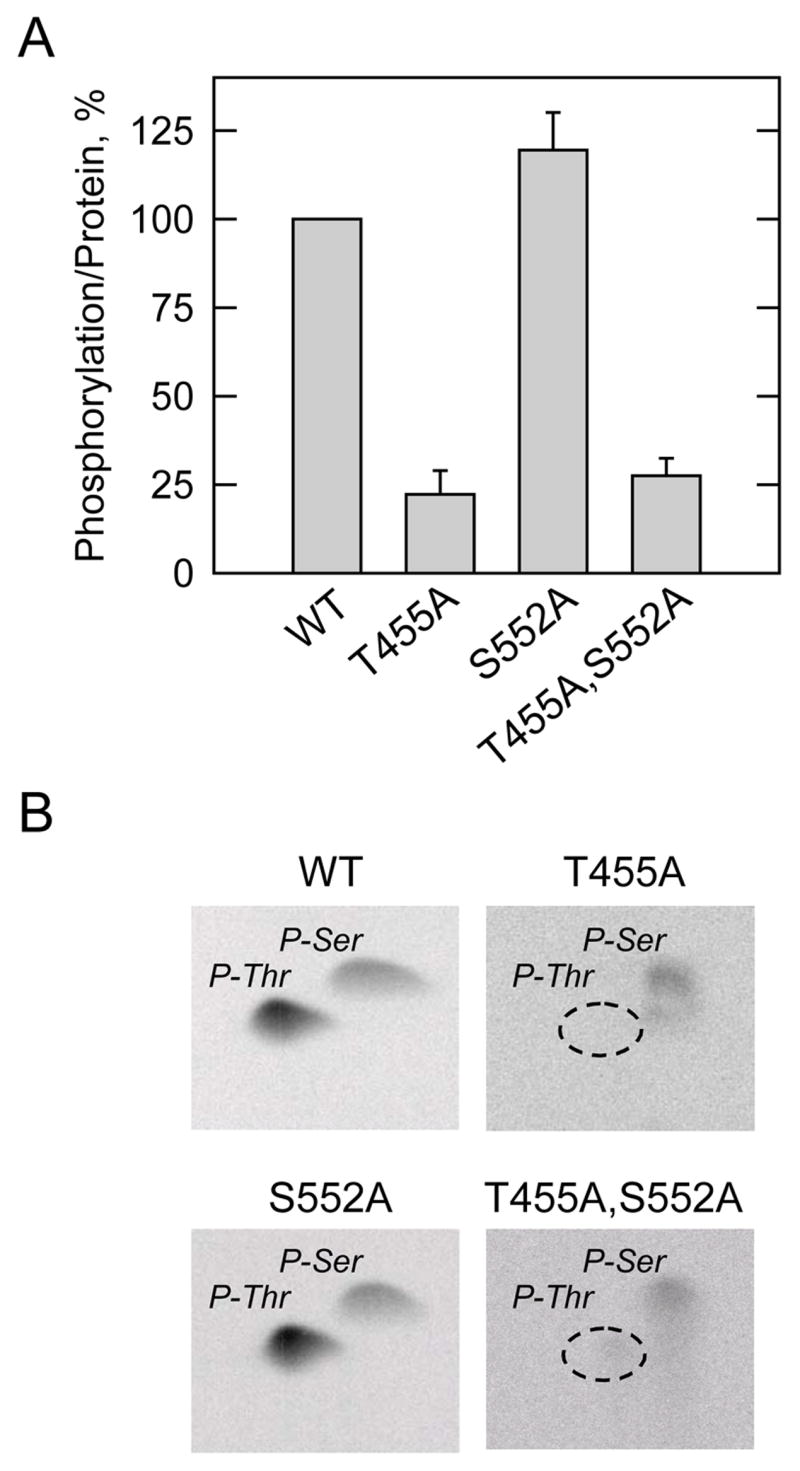
A, the wild type (WT) and the indicated mutant human CTP synthetase 1 enzymes were purified from cell extracts using Ni2+-NTA resin. The enzymes were phosphorylated with mammalian protein kinase A and [γ-32P]ATP for 10 min at 30 °C. After the phosphorylation reactions, the samples were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membrane. The phosphorylated proteins were subjected to phosphorimaging analysis and the relative amounts of phosphate incorporated were quantified using ImageQuant software. The maximum extent of wild type CTP synthetase 1 phosphorylation was set at 100%. The data were normalized to the amount of human CTP synthetase 1 proteins as determined by immunoblot analysis using anti-His6 antibodies. The values reported were average of two separate experiments ± S.D. B, the 32P-labeled wild type and indicated mutant human CTP synthetase 1 enzymes on polyvinylidene difluoride membranes were hydrolyzed with 6N HCl, and the hydrolysates were separated on cellulose thin layer plates by two-dimensional electrophoresis. The positions of the carrier standard phosphoamino acids phosphoserine (P-Ser) and phosphothreonine (P-Thr) are indicated in the figure. The position of phosphothreonine that was absent in the T455A and T455A,S552A mutant enzymes that was present in the wild type and S552A mutant enzymes is indicated in the figure. The phosphoserine signals from the T455A and T455A,S552A samples were less than the phosphoserine signals from the wild type and S552A samples because less radioactivity was applied to the cellulose thin layer plates. The data shown are representative of two independent experiments.
The effect of the T455A mutation on phosphoamino acid composition was examined following phosphorylation with protein kinase A. The T455A mutation resulted in the loss of the phosphothreonine that was present in the wild type CTP synthetase 1 (Fig. 4B). We also examined the effect of the T455A mutation on the phosphopeptide map of the human CTP synthetase 1. Following the phosphorylation with protein kinase A and 32P-labeled ATP, the wild type and mutant T455A CTP synthetase 1 enzymes were subjected to two-dimensional phosphopeptide mapping analysis (Fig. 5). The major phosphopeptide of the wild type enzyme was missing in the map of the T455A mutant enzyme. This indicated that Thr455 was contained in the major phosphopeptide present in the wild type enzyme after phosphorylation with protein kinase A. Taken together, the data supported the conclusion that Thr455 was the threonine residue in CTP synthetase 1 that was phosphorylated by protein kinase A.
FIGURE 5. Effect of the T455A mutation on the phosphopeptide map of human CTP synthetase 1 phosphorylated by protein kinase.
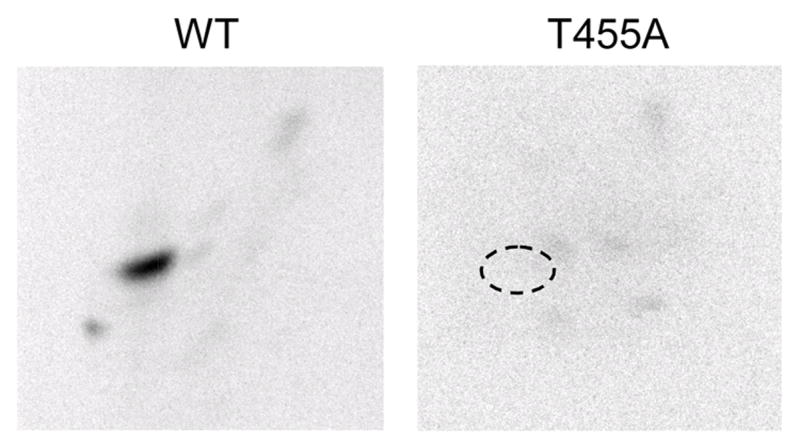
A. The purified wild type (WT) and T455A mutant human CTP synthetase 1 enzymes were phosphorylated with mammalian protein kinase A and [γ-32P]ATP for 10 min. After phosphorylation, the samples were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membrane. The 32P-labeled proteins were digested with L-1-tosylamido-2-phenylethyl chloromethyl ketone-trypsin. The resulting peptides were separated on cellulose thin layer plates by electrophoresis (from left to right) in the first dimension and by chromatography (from bottom to top) in the second dimension. The position of the major phosphopeptide that was absent in the T455A mutant enzyme that was present in the wild type enzyme is indicated in the figure. The data are representative of three independent experiments.
Although the S552 peptide was a substrate for protein kinase A phosphorylation, the S552A mutation alone, and in combination with the T455A mutation, did not affect the phosphorylation of CTP synthetase 1 (Fig. 4A). Moreover, the S552A mutation did not have a major effect on the amount of phosphoserine in the enzyme, and the phosphoamino composition of the T455A,S552A double mutant was essentially the same as that of the T455A mutant enzyme (Fig. 4B). These results indicated that Ser552 was not the serine residue within CTP synthetase 1 that was phosphorylated by protein kinase A.
Effect of the T455A Mutation on the Phosphorylation of Human CTP Synthetase 1 in S. cerevisiae
We examined the effect of the T455A mutation on the phosphorylation of CTP synthetase 1 in vivo. Exponential phase S. cerevisiae ura7Δ ura8Δ cells expressing the His6-tagged wild type and T455A alleles were labeled with 32Pi for 3h; the enzymes were isolated from cell extracts with Ni2+-NTA resin, subjected to SDS-PAGE, and transferred to PVDF membrane. Phosphorimaging analysis showed that the extent of phosphorylation of T455A mutant CTP synthetase 1 was reduced by 44% when compared with the phosphorylation of the wild type enzyme (Fig. 6A). Immunoblot analysis using anti-His6 antibodies confirmed the identification of the phosphorylated proteins. As described previously (40), the wild type human CTP synthetase 1 enzyme was primarily phosphorylated on a serine residue(s) when it was expressed in S. cerevisiae (Fig. 6B). The relative proportion of phosphoserine to phosphothreonine was about 70:30. Although the overall extent of enzyme phosphorylation was reduced in the T455A mutant enzyme (Fig. 6A), a threonine residue(s) was still phosphorylated in CTP synthetase 1 (Fig. 6B). These results indicated that CTP synthetase 1 is phosphorylated on a threonine residue(s) by a protein kinase(s) other than protein kinase A.
FIGURE 6. Effect of the T455A mutation on the phosphorylation of human CTP synthetase 1 in vivo, and phosphoamino acid analysis of phosphorylated proteins.
A, Cultures (50 ml) of S. cerevisiae ura7Δ ura8Δ cells expressing the wild type (WT) and T455A mutant human CTP synthetase 1 enzymes were grown to the exponential phase of growth. Cells were harvested and resuspended in 5 ml of fresh medium containing 32Pi (0.25 mCi/ml) and incubated for 3 h. Following the incubation, the human CTP synthetase 1 proteins were purified from cell extracts with Ni2+-NTA resin, subjected to SDS-PAGE, and transferred to polyvinylidene difluoride membrane. The 32P-labeled human CTP synthetase 1 proteins were visualized by phosphorimaging analysis, and their relative intensity was quantified using ImageQuant software. The maximum extent of wild type CTP synthetase 1 phosphorylation was set at 100%. The data were normalized to the amount of human CTP synthetase 1 proteins as determined by immunoblot analysis using anti-His6 antibodies. The data are representative of two independent experiments. B, the 32P-labeled wild type and T455A mutant human CTP synthetase 1 enzymes on polyvinylidene difluoride membranes were hydrolyzed with 6N HCl, and the hydrolysates were separated on cellulose thin layer plates by two-dimensional electrophoresis. The positions of the carrier standard phosphoamino acids phosphoserine (P-Ser) and phosphothreonine (P-Thr) are indicated in the figure. The data shown are representative of two independent experiments.
Effect of the T455A Mutation on CTP Synthetase 1 Activity In Vitro and on the Cellular Concentration of CTP In Vivo
The effect of the T455A mutation on the activity of purified human CTP synthetase 1 activity was examined. The specific activity of the T455A mutant CTP synthetase 1 was 2-fold greater than the specific activity of the purified wild type enzyme (Fig. 7A). This result supported the conclusion that phosphorylation at Thr455 by protein kinase A contributed to the inhibition of CTP synthetase 1 activity. We questioned whether the elevated CTP synthetase 1 activity of the T455A mutant enzyme would affect the cellular concentration of CTP. S. cerevisiae ura7Δ ura8Δ cells expressing the wild type and T455A mutant human CTP synthetase 1 enzymes were grown to the exponential phase of growth; nucleotides were extracted and analyzed for CTP by high performance liquid chromatography. The cellular concentration of CTP in cells that expressed the T455A mutant CTP synthetase 1 was 2.5-fold greater when compared with cells that expressed the wild type human enzyme (Fig. 7B). This result was consistent with the in vitro effect of the T455A mutation on CTP synthetase 1 activity, and supported the conclusion that phosphorylation of the enzyme at Thr455 by protein kinase A was relevant in vivo.
FIGURE 7. Effect of the T455A mutation on human CTP synthetase activity, and on the cellular concentration of CTP in S. cerevisiae.
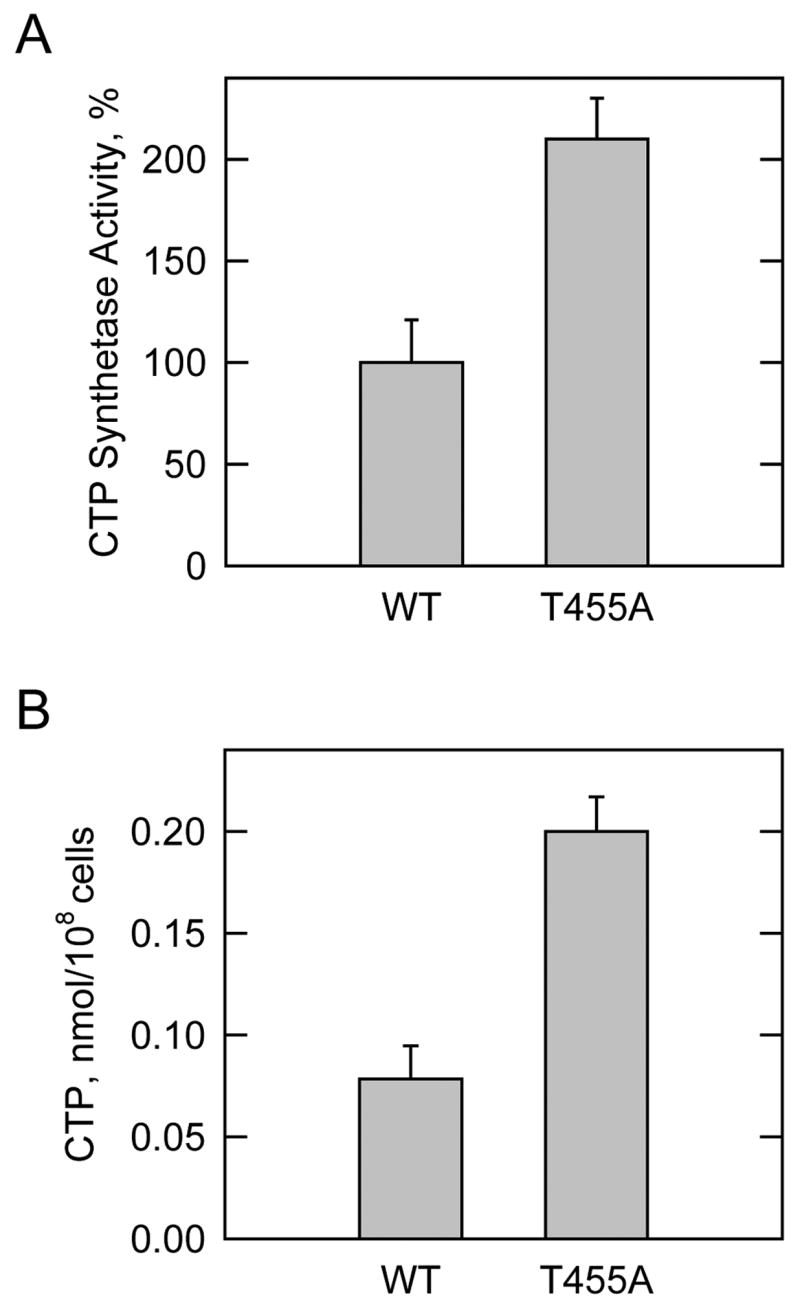
A, the wild type (WT) and T455A mutant human CTP synthetase 1 enzymes were expressed and purified from S. cerevisiae ura7Δ ura8Δ cells. CTP synthetase activity was determined with the spectrophotometric assay, and the activity of the wild type enzyme was set at 100%. B, S. cerevisiae ura7Δ ura8Δ cells expressing the wild type and T455A mutant human CTP synthetase 1 enzymes were grown to the exponential phase of growth. Nucleotides were extracted and the concentration of CTP was analyzed by high performance liquid chromatography. The values reported were average of three separate experiments ± S.D.
Effect of the T455A Mutation in CTP Synthetase 1 on Phosphatidylcholine Synthesis in S. cerevisiae
CTP plays an essential role in the synthesis of the major membrane phospholipid phosphatidylcholine in S. cerevisiae. Phosphatidylcholine is synthesized from CTP via the high energy-rich intermediates CDP-choline or CDP-diacylglycerol (74). The synthesis of phosphatidylcholine from CDP-choline is direct, whereas the synthesis of phosphatidylcholine via CDP-diacylglycerol is indirect, and occurs through the reaction sequence CDP-diacylglycerol → phosphatidylserine → phosphatidylethanolamine → → → phosphatidylcholine (74). CDP-diacylglycerol is also used directly for the synthesis of phosphatidylinositol, phosphatidylglycerol, and cardiolipin (74). Previous studies have shown that the cellular concentration of CTP, as regulated by CTP synthetase activity, affects the synthesis of phosphatidylcholine in S. cerevisiae via CDP-choline (19, 21, 75, 76). Accordingly, we questioned whether the T455A mutation in human CTP synthetase 1 affected phosphatidylcholine synthesis. In one experiment, S. cerevisiae ura7Δ ura8Δ cells expressing the wild type and T455A mutant enzymes were labeled for 30 min with 32Pi; in another experiment, the cells were labeled for 30 min with [methyl-14C]choline. The 32Pi is incorporated into phosphatidylcholine by way of CDP-diacylglycerol and by way of CDP-choline, whereas [methyl-14C]choline is only incorporated into phosphatidylcholine by way of CDP-choline (19, 21). The 32P-labeling experiment showed that the T455A mutation in CTP synthetase 1 did not have a major effect on the synthesis of phospholipids including phosphatidylcholine via CDP-diacylglycerol plus CDP-choline (Fig. 8A). On the other hand, the 14C-labeling experiment showed that the T455A mutation in the enzyme resulted in a 1.5-fold increase in the synthesis of phosphatidylcholine via CDP-choline (Fig. 8B). These data further demonstrated that the phosphorylation of CTP synthetase 1 at Thr455 was physiological relevant.
FIGURE 8. Effect of the T455A mutation on the synthesis of phospholipids.
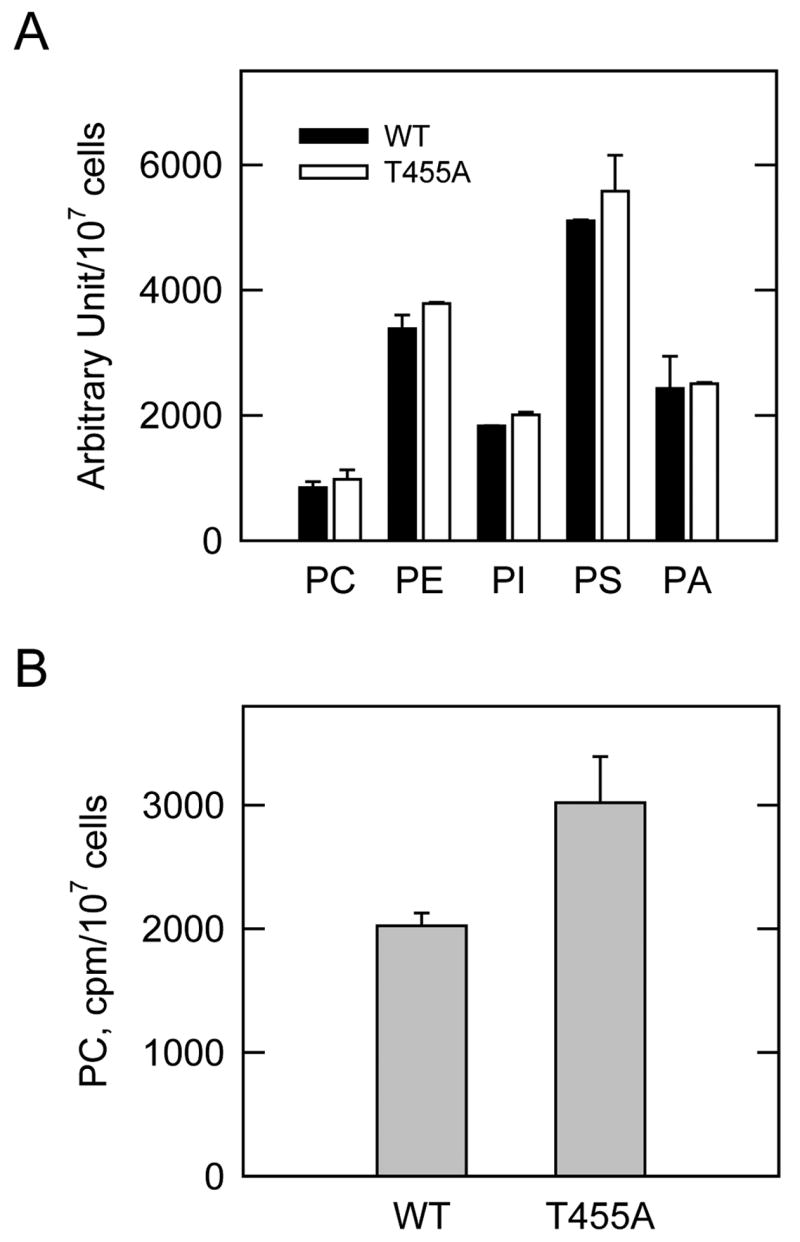
S. cerevisiae ura7Δ ura8Δ cells expressing the wild type (WT) and T455A mutant human CTP synthetase 1 enzymes were grown to the exponential phase of growth. Cells were incubated with 32Pi (5 μCi/ml) (A) or with [methyl-14C]choline (0.5 μCi/ml) (B) for 30 min to label all phospholipids and to label CDP-choline pathway-derived phosphatidylcholine, respectively. Phospholipids were extracted and analyzed by two-dimensional thin-layer chromatography. The 32P-labeled phospholipids were analyzed by phosphorimaging and quantified using ImageQuant software. The 14C-labeled phosphatidylcholine was determined by scintillation counting. Each data point represents the average of duplicate determinations from two independent experiments ± S.D.
Concluding Discussion
We initiated studies on the phosphorylation of human CTP synthetase 1 because the regulation of its activity is so important to normal cell growth and development (22–27). In this study, we demonstrated that the human enzyme expressed and purified from S. cerevisiae ura7ΔΔura8Δ cells was phosphorylated by mammalian protein kinase A, and that this phosphorylation resulted in the inhibition of CTP synthetase 1 activity. The extent of inhibition by mammalian protein kinase A phosphorylation may be an underestimate because a population of the human enzyme was already phosphorylated in yeast cells prior to its purification. The degree of this phosphorylation was not addressed. Thr455 was identified as a major site of protein kinase A phosphorylation. The in vitro and in vivo analyses of the T455A mutant CTP synthetase 1 enzyme supported the conclusion that phosphorylation at Thr455 resulted in the inhibition of CTP synthetase 1 activity. Protein kinase A also phosphorylated CTP synthetase 1 on a serine residue(s), but the identification of the specific site(s) involved will require additional studies.
The conclusion that human CTP synthetase 1 is phosphorylated by protein kinase A in S. cerevisiae cells is based on the transient increase in the extent of enzyme phosphorylation following the supplementation of glucose (a fermentable carbon source) to nonfermenting cells (40). This treatment is known to stimulate the Ras-cAMP pathway and protein kinase A activity in S. cerevisiae (77–79). The transient increase in CTP synthetase 1 phosphorylation correlates with a transient increase in the cellular concentration of CTP (40). However, the direct effect of protein kinase A phosphorylation of CTP synthetase 1 was the inhibition of its enzyme activity. The physiological consequences of glucose supplementation are much more complex than just stimulating the Ras-cAMP pathway (80). Thus, the basis for the glucose-induced transient increase in the CTP level is unclear.
The direct effect of protein kinase A phosphorylation on human CTP synthetase 1 activity was opposite to that of the S. cerevisiae URA7-encoded CTP synthetase (39, 81). The S. cerevisiae enzyme is phosphorylated on Ser424, and this phosphorylation results in the stimulation of CTP synthetase activity (81). The protein kinase A phosphorylation sites in the yeast (i.e., Ser424) and the human (i.e., Thr455) enzymes are not conserved. The opposing effects of protein kinase A phosphorylation on the CTP synthetase enzymes from yeast and human may reflect differences in the regulatory functions of protein kinase A in these organisms. In S. cerevisiae, protein kinase A is the enzyme through which the Ras-cAMP pathway mediates its signal for cell growth (77–79). Activation of Ras and the subsequent elevation of protein kinase A activity by cAMP production are associated with rapid cell growth and enhanced metabolic activity of the cell (77–79). Stimulation of CTP synthetase activity by protein kinase A phosphorylation may provide a mechanism by which the Ras-cAMP pathway mediates the synthesis of the CTP required for yeast cell growth. In mammalian cells, cAMP levels and protein kinase A activity are not controlled through the Ras pathway, and the roles of protein kinase A activity in cell physiology are much more complex than in the single cell yeast (82). The inhibition of CTP synthetase 1 activity in response to protein kinase A phosphorylation may be a mechanism by which CTP synthesis and cell growth are attenuated in specific tissues/organs in humans. The knowledge that CTP synthetase 1 is phosphorylated by protein kinase A at Thr455 provides a basis for future studies to examine protein kinase A phosphorylation and regulation of CTP synthetase 1 in specific human cell types.
TABLE 1.
Strains used in this work
| Strain | Relevant characteristics | Source or Ref. |
|---|---|---|
| E. coli | ||
| DH5α | F−, φ80dlacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rk− mk+), phoA, supE44, λ−thi-1, gyrA96, relA1 | (53) |
| S. cerevisiae | ||
| SDO195 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO134] | (19) |
| GHY55 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1] | (40) |
| MCY24 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1(T83A)] | This study |
| MCY20 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1(S219A)] | This study |
| MCY25 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1(T378A)] | This study |
| MCY26 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1(T455A)] | This study |
| MCY21 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1(S552A)] | This study |
| MCY22 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1(S562A)] | This study |
| MCY27 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1(T566A)] | This study |
| MCY23 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1(S573A)] | This study |
| MCY28 | MATa leu2-3,112 trp1-289 ura3-52 ura7Δ::TRP1 ura8Δ::hisG [pDO105-hCTPS1(T455A,S552A)] | This study |
Acknowledgments
We thank Gil-Soo Han and Avula Sreenivas for their critical review of this manuscript.
Footnotes
This work was supported in part by United States Public Health Service, National Institutes of Health Grant GM-50679.
The abbreviations used are: PVDF, polyvinylidene difluoride
References
- 1.Liberman I. J Biol Chem. 1956;222:765–775. [PubMed] [Google Scholar]
- 2.Long CW, Pardee AB. J Biol Chem. 1967;242:4715–4721. [PubMed] [Google Scholar]
- 3.Levitzki A, Koshland DE., Jr Biochemistry. 1972;11:241–246. doi: 10.1021/bi00752a015. [DOI] [PubMed] [Google Scholar]
- 4.Levitzki A, Koshland DE., Jr Proc Nat Acad Sci USA. 1969;62:1121–1128. doi: 10.1073/pnas.62.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levitzki A, Koshland DE., Jr Biochemistry. 1971;10:3365–3371. doi: 10.1021/bi00794a008. [DOI] [PubMed] [Google Scholar]
- 6.Lewis DA, Villafranca JJ. Biochemistry. 1989;28:8454–8459. doi: 10.1021/bi00447a027. [DOI] [PubMed] [Google Scholar]
- 7.von der Saal W, Anderson PM, Villafranca JJ. J Biol Chem. 1985;260:14993–14997. [PubMed] [Google Scholar]
- 8.Yang WL, McDonough VM, Ozier-Kalogeropoulos O, Adeline MT, Flocco MT, Carman GM. Biochemistry. 1994;33:10785–10793. doi: 10.1021/bi00201a028. [DOI] [PubMed] [Google Scholar]
- 9.Nadkarni AK, McDonough VM, Yang WL, Stukey JE, Ozier-Kalogeropoulos O, Carman GM. J Biol Chem. 1995;270:24982–24988. doi: 10.1074/jbc.270.42.24982. [DOI] [PubMed] [Google Scholar]
- 10.Wadskov-Hansen SL, Willemoes M, Martinussen J, Hammer K, Neuhard J, Larsen S. J Biol Chem. 2001;276:38002–38009. doi: 10.1074/jbc.M100531200. [DOI] [PubMed] [Google Scholar]
- 11.Levitzki A, Koshland DE., Jr Biochemistry. 1972;11:247–252. doi: 10.1021/bi00752a016. [DOI] [PubMed] [Google Scholar]
- 12.Pappas A, Yang WL, Park TS, Carman GM. J Biol Chem. 1998;273:15954–15960. doi: 10.1074/jbc.273.26.15954. [DOI] [PubMed] [Google Scholar]
- 13.Stryer L. Biochemistry. 4. W.H. Freeman and Company; New York: 1995. [Google Scholar]
- 14.Aronow B, Ullman B. J Biol Chem. 1987;262:5106–5112. [PubMed] [Google Scholar]
- 15.Robert de Saint Vincent B, Buttin G. Biochim Biophys Acta. 1980;610:352–359. [Google Scholar]
- 16.Meuth M, L’Heureux-Huard N, Trudel M. Proc Nat Acad Sci USA. 1979;76:6505–6509. doi: 10.1073/pnas.76.12.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozier-Kalogeropoulos O, Fasiolo F, Adeline MT, Collin J, Lacroute F. Mol Gen Genet. 1991;231:7–16. doi: 10.1007/BF00293815. [DOI] [PubMed] [Google Scholar]
- 18.Ozier-Kalogeropoulos O, Adeline MT, Yang WL, Carman GM, Lacroute F. Mol Gen Genet. 1994;242:431–439. doi: 10.1007/BF00281793. [DOI] [PubMed] [Google Scholar]
- 19.Ostrander DB, O’Brien DJ, Gorman JA, Carman GM. J Biol Chem. 1998;273:18992–19001. doi: 10.1074/jbc.273.30.18992. [DOI] [PubMed] [Google Scholar]
- 20.Hatch GM, McClarty G. J Biol Chem. 1996;271:25810–25816. doi: 10.1074/jbc.271.42.25810. [DOI] [PubMed] [Google Scholar]
- 21.McDonough VM, Buxeda RJ, Bruno MEC, Ozier-Kalogeropoulos O, Adeline MT, McMaster CR, Bell RM, Carman GM. J Biol Chem. 1995;270:18774–18780. doi: 10.1074/jbc.270.32.18774. [DOI] [PubMed] [Google Scholar]
- 22.van den Berg AA, van Lenthe H, Busch S, de Korte D, Roos D, van Kuilenburg ABP, van Gennip AH. Eur J Biochem. 1993;216:161–167. doi: 10.1111/j.1432-1033.1993.tb18128.x. [DOI] [PubMed] [Google Scholar]
- 23.van den Berg AA, van Lenthe H, Kipp JB, de Korte D, Van Kuilenburg AB, van Gennip AH. Eur J Cancer. 1995;31A:108–112. doi: 10.1016/0959-8049(94)00442-8. [DOI] [PubMed] [Google Scholar]
- 24.Verschuur AC, van Gennip AH, Muller EJ, Voute PA, Van Kuilenburg AB. Adv Exp Med Biol. 1998;431:667–671. doi: 10.1007/978-1-4615-5381-6_129. [DOI] [PubMed] [Google Scholar]
- 25.Kizaki H, Williams JC, Morris HP, Weber G. Cancer Res. 1980;40:3921–3927. [PubMed] [Google Scholar]
- 26.Weber G, Lui MS, Takeda E, Denton JE. Life Sci. 1980;27:793–799. doi: 10.1016/0024-3205(80)90333-1. [DOI] [PubMed] [Google Scholar]
- 27.Weber G, Olah E, Lui MS, Tzeng D. Adv Enzyme Regul. 1979;17:1–21. doi: 10.1016/0065-2571(79)90005-0. [DOI] [PubMed] [Google Scholar]
- 28.Hindenburg AA, Taub RN, Grant S, Chang G, Baker MA. Cancer Res. 1985;45:3048–3052. [PubMed] [Google Scholar]
- 29.Kang GJ, Cooney DA, Moyer JD, Kelley JA, Kim HY, Marquez VE, Johns DG. J Biol Chem. 1989;264:713–718. [PubMed] [Google Scholar]
- 30.Politi PM, Xie F, Dahut W, Ford H, Jr, Kelley JA, Bastian A, Setser A, Allegra CJ, Chen AP, Hamilton JM. Cancer Chemother Pharmacol. 1995;36:513–523. doi: 10.1007/BF00685802. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Cooney DA, Zhang MH, Ahluwalia G, Ford H, Jr, Johns DG. Cancer Res. 1993;53:5714–5720. [PubMed] [Google Scholar]
- 32.Verschuur AC, van Gennip AH, Leen R, Meinsma R, Voute PA, van Kuilenburg ABP. Br J Haematol. 2000;110:161–169. doi: 10.1046/j.1365-2141.2000.02136.x. [DOI] [PubMed] [Google Scholar]
- 33.Verschuur AC, van Gennip AH, Leen R, Meinsma R, Voute PA, Van Kuilenburg AB. Br J Haematol. 2000;110:161–169. doi: 10.1046/j.1365-2141.2000.02136.x. [DOI] [PubMed] [Google Scholar]
- 34.Verschuur AC, van Gennip AH, Leen R, Muller EJ, Elzinga L, Voute PA, Van Kuilenburg AB. Eur J Cancer. 2000;36:627–635. doi: 10.1016/s0959-8049(00)00021-6. [DOI] [PubMed] [Google Scholar]
- 35.Hofer A, Steverding D, Chabes A, Brun R, Thelander L. Proc Natl Acad Sci USA. 2001;98:6412–6416. doi: 10.1073/pnas.111139498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dereuddre-Bosquet N, Roy B, Routledge K, Clayette P, Foucault G, Lepoivre M. Antiviral Res. 2004;61:67–70. doi: 10.1016/j.antiviral.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Thomas PE, Lamb BJ, Chu EHY. Biochim Biophys Acta. 1988;953:334–344. doi: 10.1016/0167-4838(88)90042-8. [DOI] [PubMed] [Google Scholar]
- 38.Yang WL, Bruno MEC, Carman GM. J Biol Chem. 1996;271:11113–11119. doi: 10.1074/jbc.271.19.11113. [DOI] [PubMed] [Google Scholar]
- 39.Yang WL, Carman GM. J Biol Chem. 1996;271:28777–28783. doi: 10.1074/jbc.271.46.28777. [DOI] [PubMed] [Google Scholar]
- 40.Han GS, Sreenivas A, Choi MG, Chang YF, Martin SS, Baldwin EP, Carman GM. J Biol Chem. 2005;280:38328–38336. doi: 10.1074/jbc.M509622200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trudel M, van Genechten T, Meuth M. J Biol Chem. 1984;259:2355–2359. [PubMed] [Google Scholar]
- 42.Meuth M, Goncalves O, Thom P. Somat Cell Genet. 1982;8:423–432. doi: 10.1007/BF01538705. [DOI] [PubMed] [Google Scholar]
- 43.Aronow B, Watts T, Lassetter J, Washtien W, Ullman B. J Biol Chem. 1984;259:9035–9043. [PubMed] [Google Scholar]
- 44.Kaufman ER. Muta Res. 1986;161:19–27. doi: 10.1016/0027-5107(86)90096-5. [DOI] [PubMed] [Google Scholar]
- 45.Chu EHY, McLaren JD, Li IC, Lamb B. Biochem Genet. 1984;22:701–715. doi: 10.1007/BF00485854. [DOI] [PubMed] [Google Scholar]
- 46.Yang WL, Carman GM. J Biol Chem. 1995;270:14983–14988. doi: 10.1074/jbc.270.25.14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamauchi M, Yamauchi N, Meuth M. EMBO J. 1990;9:2095–2099. doi: 10.1002/j.1460-2075.1990.tb07377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Kuilenburg AB, Meinsma R, Vreken P, Waterham HR, van Gennip AH. Adv Exp Med Biol. 2000;486:257–261. doi: 10.1007/0-306-46843-3_50. [DOI] [PubMed] [Google Scholar]
- 49.Endrizzi JA, Kim H, Anderson PM, Baldwin EP. Biochemistry. 2004;43:6447–6463. doi: 10.1021/bi0496945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goto M, Omi R, Nakagawa N, Miyahara I, Hirotsu K. Structure (Camb) 2004;12:1413–1423. doi: 10.1016/j.str.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 51.Kursula P, Flodin S, Ehn M, Hammarstrom M, Schuler H, Nordlund P, Stenmark P. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2006;62:613–617. doi: 10.1107/S1744309106018136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose MD, Winston F, Heiter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1990. [Google Scholar]
- 53.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. 2. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1989. [Google Scholar]
- 54.Fernandez F, Rush JS, Toke DA, Han GS, Quinn JE, Carman GM, Choi JY, Voelker DR, Aebi M, Waechter CJ. J Biol Chem. 2001;276:41455–41464. doi: 10.1074/jbc.M105544200. [DOI] [PubMed] [Google Scholar]
- 55.Ito H, Yasuki F, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schiestl RH, Gietz RD. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 57.Innis MA, Gelfand DH. In: PCR Protocols. A Guide to Methods and Applications. Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. Academic Press, Inc; San Diego: 1990. pp. 3–12. [Google Scholar]
- 58.Sikorski RS, Boeke JD. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 59.Mole ML, Hunter DL, Gao P, Lau C. Anal Biochem. 1998;259:245–252. doi: 10.1006/abio.1998.2647. [DOI] [PubMed] [Google Scholar]
- 60.Pappas A, Park TS, Carman GM. Biochemistry. 1999;38:16671–16677. doi: 10.1021/bi9920127. [DOI] [PubMed] [Google Scholar]
- 61.Laemmli UK. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 62.Haid A, Suissa M. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- 63.Boyle WJ, Van der Geer P, Hunter T. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 64.MacDonald JIS, Kent C. J Biol Chem. 1994;269:10529–10537. [PubMed] [Google Scholar]
- 65.Atkinson K, Fogel S, Henry SA. J Biol Chem. 1980;255:6653–6661. [PubMed] [Google Scholar]
- 66.Atkinson KD, Jensen B, Kolat AI, Storm EM, Henry SA, Fogel S. J Bacteriol. 1980;141:558–564. doi: 10.1128/jb.141.2.558-564.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bligh EG, Dyer WJ. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 68.Han GS, Johnston CN, Carman GM. J Biol Chem. 2004;279:5338–5345. doi: 10.1074/jbc.M311779200. [DOI] [PubMed] [Google Scholar]
- 69.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krebs EG, Beavo JA. Ann Rev Biochem. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- 71.Chock PB, Rhee SG, Stadtman ER. Annu Rev Biochem. 1980;49:813–843. doi: 10.1146/annurev.bi.49.070180.004121. [DOI] [PubMed] [Google Scholar]
- 72.Skalhegg BS, Tasken K. Front Biosci. 2000;5:D678–D693. doi: 10.2741/skalhegg. [DOI] [PubMed] [Google Scholar]
- 73.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 74.Carman GM, Henry SA. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 75.Park TS, O’Brien DJ, Carman GM. J Biol Chem. 2003;278:20785–20794. doi: 10.1074/jbc.M301394200. [DOI] [PubMed] [Google Scholar]
- 76.Choi MG, Park TS, Carman GM. J Biol Chem. 2003;278:23610–23616. doi: 10.1074/jbc.M303337200. [DOI] [PubMed] [Google Scholar]
- 77.Thevelein JM, Beullens M. J Gen Microbiol. 1985;131:3199–3209. doi: 10.1099/00221287-131-12-3199. [DOI] [PubMed] [Google Scholar]
- 78.Thevelein JM. Yeast. 1994;10:1753–1790. doi: 10.1002/yea.320101308. [DOI] [PubMed] [Google Scholar]
- 79.Broach JR, Deschenes RJ. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- 80.Fraenkel DG. In: The molecular biology of the yeast Saccharomyces. Metabolism and gene expression. Strathern JN, Jones EW, Broach JR, editors. Cold Spring Harbor Laboratory; Cold Spring Harbor: 1982. pp. 1–37. [Google Scholar]
- 81.Park TS, Ostrander DB, Pappas A, Carman GM. Biochemistry. 1999;38:8839–8848. doi: 10.1021/bi990784x. [DOI] [PubMed] [Google Scholar]
- 82.Krauss G. Biochemistry of signal transduction and regulation. 3. Wiley-VCH, Weinheim; Germany: 2003. [Google Scholar]
- 83.Gietz RD, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]