Abstract
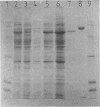
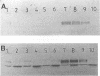
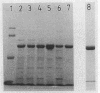
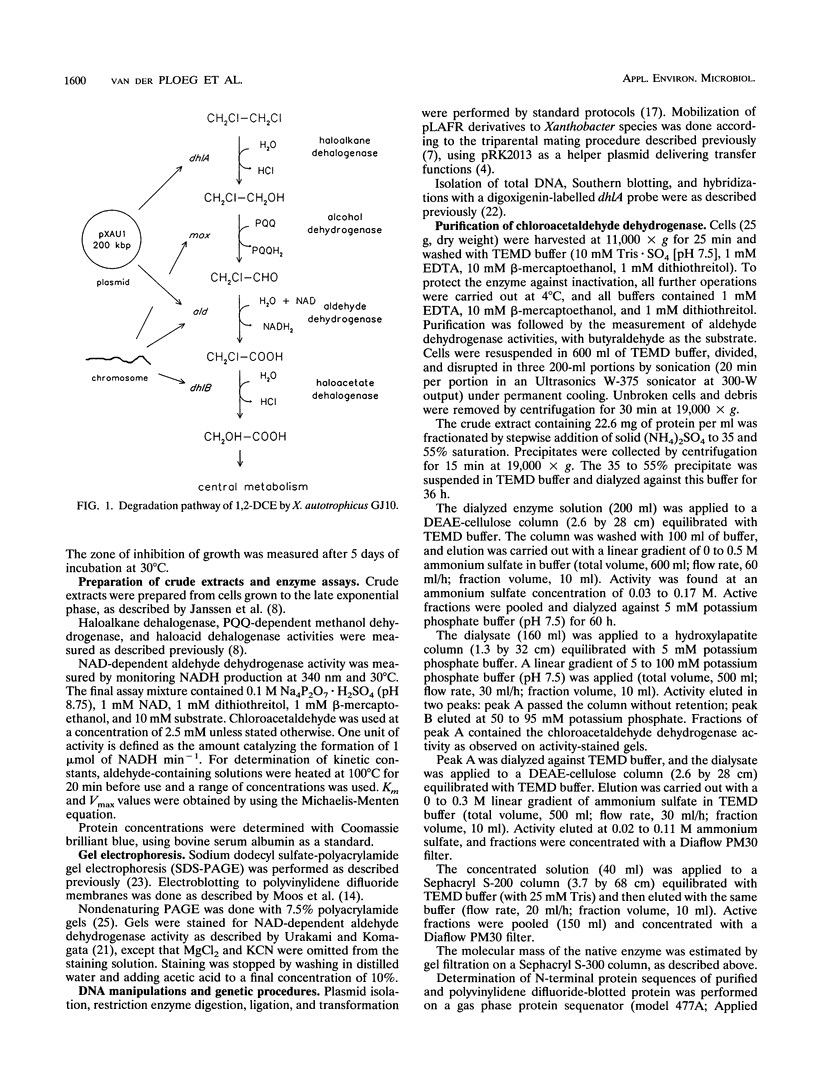
The degradation of 1,2-dichloroethane and 2-chloroethanol by Xanthobacter autotrophicus GJ10 proceeds via chloroacetaldehyde, a reactive and potentially toxic intermediate. The organism produced at least three different aldehyde dehydrogenases, of which one is plasmid encoded. Two mutants of strain GJ10, designated GJ10M30 and GJ10M41, could no longer grow on 2-chloroethanol and were found to lack the NAD-dependent aldehyde dehydrogenase that is the predominant protein in wild-type cells growing on 2-chloroethanol. Mutant GJ10M30, selected on the basis of its resistance to 1,2-dibromoethane, also had lost haloalkane dehalogenase activity and Hg2+ resistance, indicating plasmid loss. From a gene bank of strain GJ10, different clones that complemented one of these mutants were isolated. In both transconjugants, the aldehyde dehydrogenase that was absent in the mutants was overexpressed. The enzyme was purified and was a tetrameric protein of 55-kDa subunits. The substrate range was rather broad, with the highest activity measured for acetaldehyde. The Km value for chloroacetaldehyde was 160 μM, higher than those for other aldehydes tested. It is concluded that the ability of GJ10 to grow with 2-chloroethanol is due to the high expression level of an aldehyde dehydrogenase with a rather low activity for chloroacetaldehyde.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Eckfeldt J. H., Yonetani T. Isozymes of aldehyde dehydrogenase from horse liver. Methods Enzymol. 1982;89(Pt 500):474–479. doi: 10.1016/s0076-6879(82)89081-2. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen D. B., Pries F., van der Ploeg J., Kazemier B., Terpstra P., Witholt B. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J Bacteriol. 1989 Dec;171(12):6791–6799. doi: 10.1128/jb.171.12.6791-6799.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen D. B., Scheper A., Dijkhuizen L., Witholt B. Degradation of halogenated aliphatic compounds by Xanthobacter autotrophicus GJ10. Appl Environ Microbiol. 1985 Mar;49(3):673–677. doi: 10.1128/aem.49.3.673-677.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrossek D., Steinbüchel A., Schlegel H. G. Three different proteins exhibiting NAD-dependent acetaldehyde dehydrogenase activity from Alcaligenes eutrophus. Eur J Biochem. 1987 Sep 15;167(3):541–548. doi: 10.1111/j.1432-1033.1987.tb13371.x. [DOI] [PubMed] [Google Scholar]
- Keuning S., Janssen D. B., Witholt B. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol. 1985 Aug;163(2):635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok M., Oldenhuis R., van der Linden M. P., Meulenberg C. H., Kingma J., Witholt B. The Pseudomonas oleovorans alkBAC operon encodes two structurally related rubredoxins and an aldehyde dehydrogenase. J Biol Chem. 1989 Apr 5;264(10):5442–5451. [PubMed] [Google Scholar]
- McCann J., Simmon V., Streitwieser D., Ames B. N. Mutagenicity of chloroacetaldehyde, a possible metabolic product of 1,2-dichloroethane (ethylene dichloride), chloroethanol (ethylene chlorohydrin), vinyl chloride, and cyclophosphamide. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3190–3193. doi: 10.1073/pnas.72.8.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moos M., Jr, Nguyen N. Y., Liu T. Y. Reproducible high yield sequencing of proteins electrophoretically separated and transferred to an inert support. J Biol Chem. 1988 May 5;263(13):6005–6008. [PubMed] [Google Scholar]
- Priefert H., Krüger N., Jendrossek D., Schmidt B., Steinbüchel A. Identification and molecular characterization of the gene coding for acetaldehyde dehydrogenase II (acoD) of Alcaligenes eutrophus. J Bacteriol. 1992 Feb;174(3):899–907. doi: 10.1128/jb.174.3.899-907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigal D., Cunningham S. J., Farrés J., Weiner H. Molecular cloning of the mitochondrial aldehyde dehydrogenase gene of Saccharomyces cerevisiae by genetic complementation. J Bacteriol. 1991 May;173(10):3199–3208. doi: 10.1128/jb.173.10.3199-3208.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif G., Greer C. W., Labbé D., Lau P. C. Involvement of a large plasmid in the degradation of 1,2-dichloroethane by Xanthobacter autotrophicus. Appl Environ Microbiol. 1991 Jun;57(6):1853–1857. doi: 10.1128/aem.57.6.1853-1857.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver C. A., Lidstrom M. E. Methanol dissimilation in Xanthobacter H4-14: activities, induction and comparison to Pseudomonas AM1 and Paracoccus denitrificans. J Gen Microbiol. 1985 Sep;131(9):2183–2197. doi: 10.1099/00221287-131-9-2183. [DOI] [PubMed] [Google Scholar]
- van den Wijngaard A. J., van der Kamp K. W., van der Ploeg J., Pries F., Kazemier B., Janssen D. B. Degradation of 1,2-dichloroethane by Ancylobacter aquaticus and other facultative methylotrophs. Appl Environ Microbiol. 1992 Mar;58(3):976–983. doi: 10.1128/aem.58.3.976-983.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg J., van Hall G., Janssen D. B. Characterization of the haloacid dehalogenase from Xanthobacter autotrophicus GJ10 and sequencing of the dhlB gene. J Bacteriol. 1991 Dec;173(24):7925–7933. doi: 10.1128/jb.173.24.7925-7933.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]