Abstract
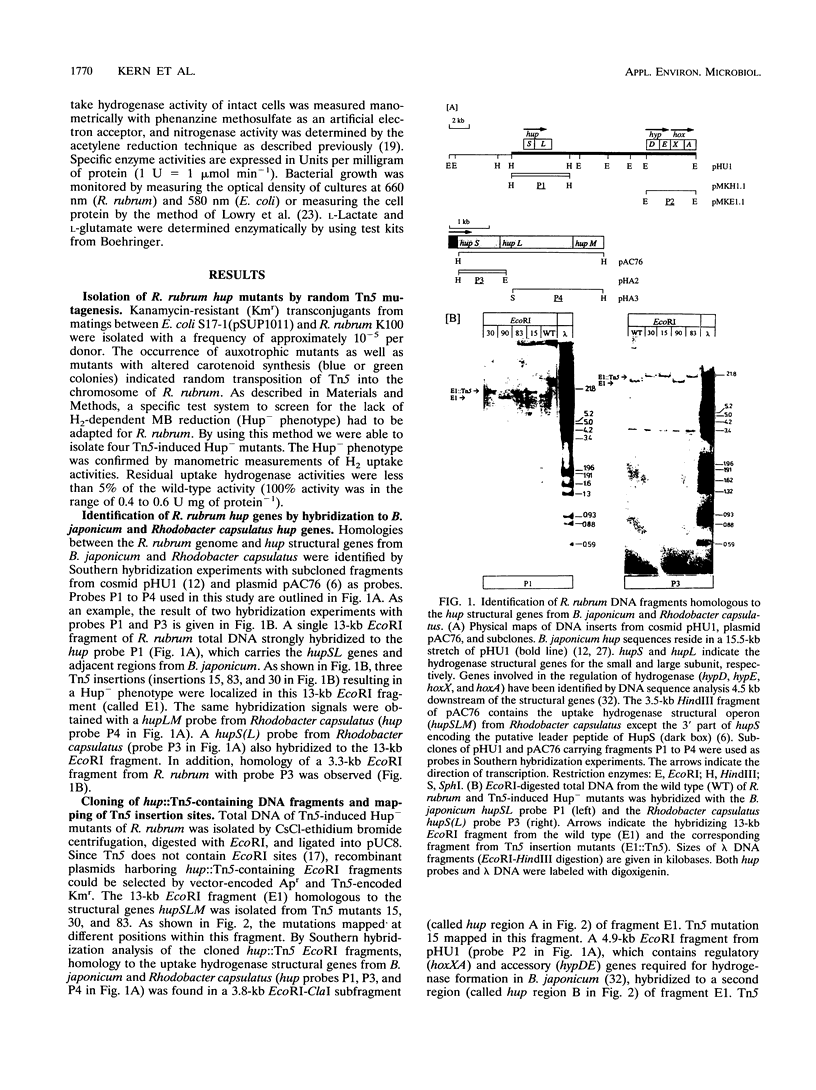
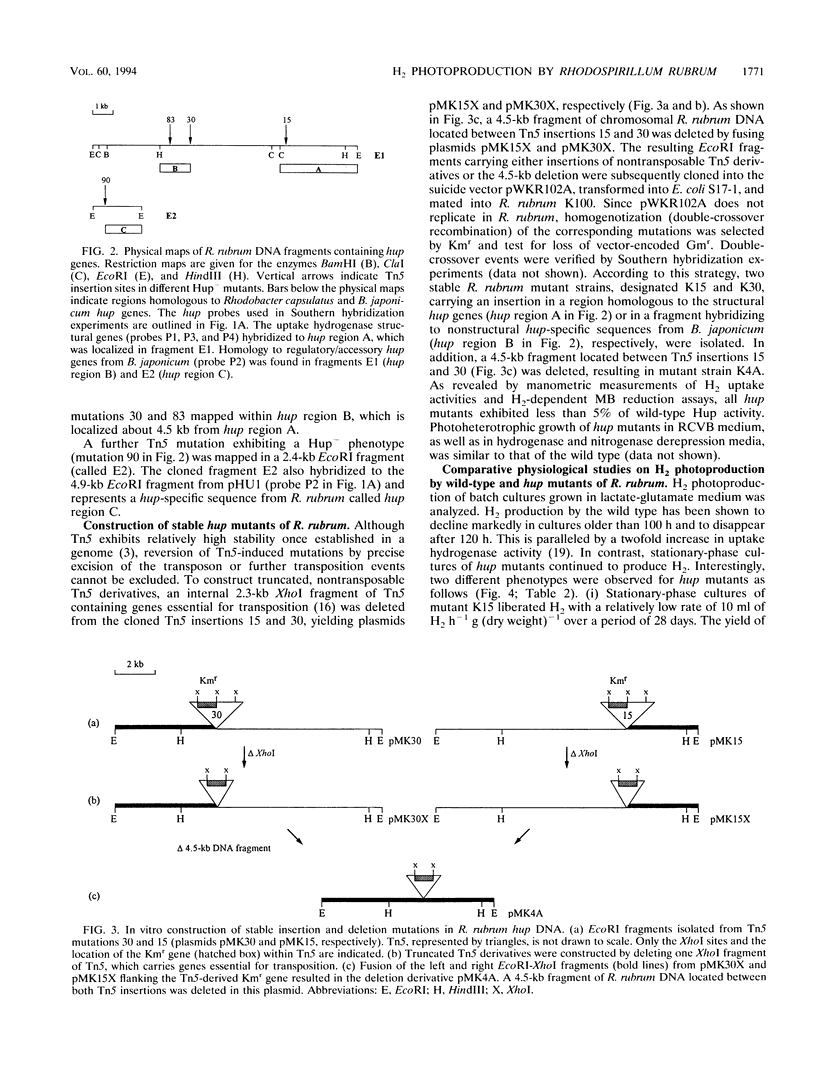
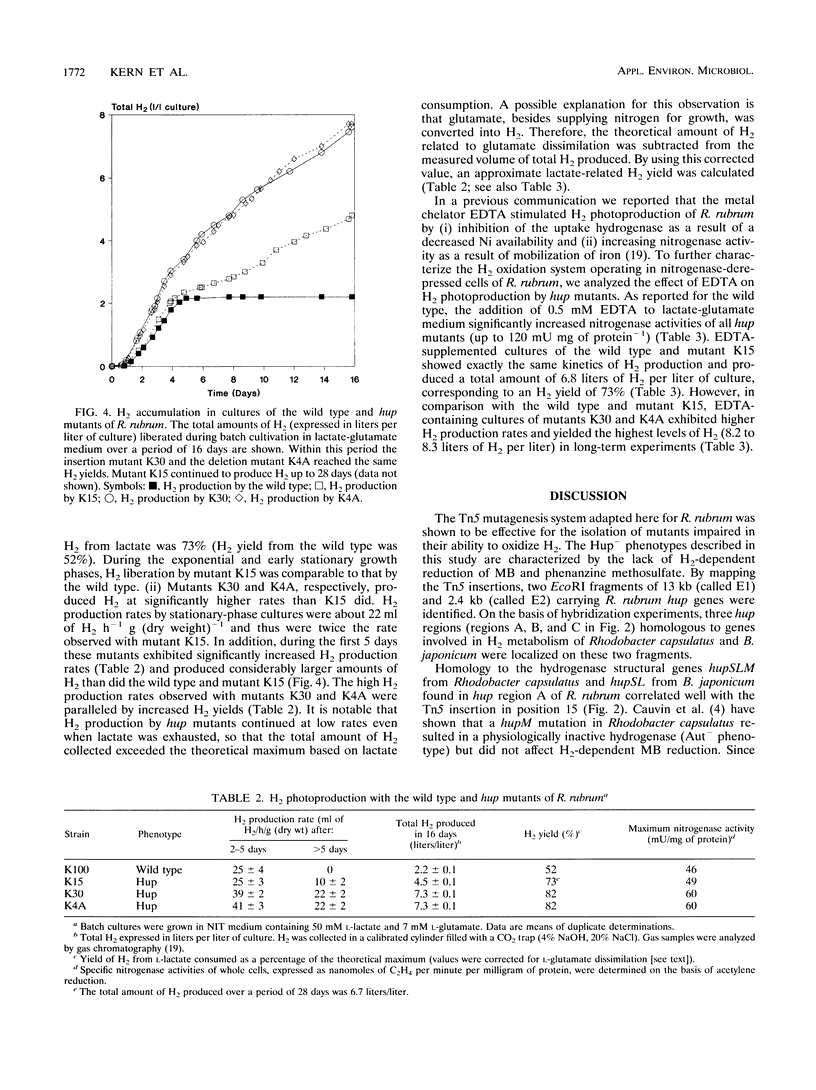
Transposon Tn5 mutagenesis was used to isolate mutants of Rhodospirillum rubrum which lack uptake hydrogenase (Hup) activity. Three Tn5 insertions mapped at different positions within the same 13-kb EcoRI fragment (fragment E1). Hybridization experiments revealed homology to the structural hydrogenase genes hupSLM from Rhodobacter capsulatus and hupSL from Bradyrhizobium japonicum in a 3.8-kb EcoRI-ClaI subfragment of fragment E1. It is suggested that this region contains at least some of the structural genes encoding the nickel-dependent uptake hydrogenase of R. rubrum. At a distance of about 4.5 kb from the fragment homologous to hupSLM, a region with homology to a DNA fragment carrying hypDE and hoxXA from B. japonicum was identified. Stable insertion and deletion mutations were generated in vitro and introduced into R. rubrum by homogenotization. In comparison with the wild type, the resulting hup mutants showed increased nitrogenase-dependent H2 photoproduction. However, a mutation in a structural hup gene did not result in maximum H2 production rates, indicating that the capacity to recycle H2 was not completely lost. Highest H2 production rates were obtained with a mutant carrying an insertion in a nonstructural hup-specific sequence and with a deletion mutant affected in both structural and nonstructural hup genes. Thus, besides the known Hup activity, a second, previously unknown Hup activity seems to be involved in H2 recycling. A single regulatory or accessory gene might be responsible for both enzymes. In contrast to the nickel-dependent uptake hydrogenase, the second Hup activity seems to be resistant to the metal chelator EDTA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. W. The structure and mechanism of iron-hydrogenases. Biochim Biophys Acta. 1990 Nov 5;1020(2):115–145. doi: 10.1016/0005-2728(90)90044-5. [DOI] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Cauvin B., Colbeau A., Vignais P. M. The hydrogenase structural operon in Rhodobacter capsulatus contains a third gene, hupM, necessary for the formation of a physiologically competent hydrogenase. Mol Microbiol. 1991 Oct;5(10):2519–2527. doi: 10.1111/j.1365-2958.1991.tb02098.x. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbeau A., Richaud P., Toussaint B., Caballero F. J., Elster C., Delphin C., Smith R. L., Chabert J., Vignais P. M. Organization of the genes necessary for hydrogenase expression in Rhodobacter capsulatus. Sequence analysis and identification of two hyp regulatory mutants. Mol Microbiol. 1993 Apr;8(1):15–29. doi: 10.1111/j.1365-2958.1993.tb01199.x. [DOI] [PubMed] [Google Scholar]
- Colonna-Romano S., Arnold W., Schlüter A., Boistard P., Pühler A., Priefer U. B. An Fnr-like protein encoded in Rhizobium leguminosarum biovar viciae shows structural and functional homology to Rhizobium meliloti FixK. Mol Gen Genet. 1990 Aug;223(1):138–147. doi: 10.1007/BF00315806. [DOI] [PubMed] [Google Scholar]
- Eberz G., Friedrich B. Three trans-acting regulatory functions control hydrogenase synthesis in Alcaligenes eutrophus. J Bacteriol. 1991 Mar;173(6):1845–1854. doi: 10.1128/jb.173.6.1845-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauque G., Peck H. D., Jr, Moura J. J., Huynh B. H., Berlier Y., DerVartanian D. V., Teixeira M., Przybyla A. E., Lespinat P. A., Moura I. The three classes of hydrogenases from sulfate-reducing bacteria of the genus Desulfovibrio. FEMS Microbiol Rev. 1988 Dec;4(4):299–344. doi: 10.1111/j.1574-6968.1988.tb02748.x. [DOI] [PubMed] [Google Scholar]
- Friedrich B., Schwartz E. Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu Rev Microbiol. 1993;47:351–383. doi: 10.1146/annurev.mi.47.100193.002031. [DOI] [PubMed] [Google Scholar]
- Haugland R. A., Cantrell M. A., Beaty J. S., Hanus F. J., Russell S. A., Evans H. J. Characterization of Rhizobium japonicum hydrogen uptake genes. J Bacteriol. 1984 Sep;159(3):1006–1012. doi: 10.1128/jb.159.3.1006-1012.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Jacobi A., Rossmann R., Böck A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158(6):444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- Jahn A., Keuntje B., Dörffler M., Klipp W., Oelze J. Optimizing photoheterotrophic H2 production by Rhodobacter capsulatus upon interposon mutagenesis in the hupL gene. Appl Microbiol Biotechnol. 1994 Jan;40(5):687–690. doi: 10.1007/BF00173330. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Yin J. C., Reznikoff W. S. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell. 1982 Oct;30(3):873–882. doi: 10.1016/0092-8674(82)90292-6. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kerby R. L., Hong S. S., Ensign S. A., Coppoc L. J., Ludden P. W., Roberts G. P. Genetic and physiological characterization of the Rhodospirillum rubrum carbon monoxide dehydrogenase system. J Bacteriol. 1992 Aug;174(16):5284–5294. doi: 10.1128/jb.174.16.5284-5294.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp W., Masepohl B., Pühler A. Identification and mapping of nitrogen fixation genes of Rhodobacter capsulatus: duplication of a nifA-nifB region. J Bacteriol. 1988 Feb;170(2):693–699. doi: 10.1128/jb.170.2.693-699.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Morrison D. L., Magargal L. E., Ehrlich D. R., Goldberg R. E., Robb-Doyle E. Review of choroidal osteoma: successful krypton red laser photocoagulation of an associated subretinal neovascular membrane involving the fovea. Ophthalmic Surg. 1987 Apr;18(4):299–303. [PubMed] [Google Scholar]
- Sayavedra-Soto L. A., Powell G. K., Evans H. J., Morris R. O. Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8395–8399. doi: 10.1073/pnas.85.22.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M. B., Orlova E. V., Smirnova E. A., Hovmöller S., Zorin N. A. Three-dimensional structure of the nickel-containing hydrogenase from Thiocapsa roseopersicina. J Bacteriol. 1991 Apr;173(8):2576–2580. doi: 10.1128/jb.173.8.2576-2580.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soom C., Verreth C., Sampaio M. J., Vanderleyden J. Identification of a potential transcriptional regulator of hydrogenase activity in free-living Bradyrhizobium japonicum strains. Mol Gen Genet. 1993 May;239(1-2):235–240. doi: 10.1007/BF00281623. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vignais P. M., Colbeau A., Willison J. C., Jouanneau Y. Hydrogenase, nitrogenase, and hydrogen metabolism in the photosynthetic bacteria. Adv Microb Physiol. 1985;26:155–234. doi: 10.1016/s0065-2911(08)60397-5. [DOI] [PubMed] [Google Scholar]
- Vignais P. M., Toussaint B. Molecular biology of membrane-bound H2 uptake hydrogenases. Arch Microbiol. 1994;161(1):1–10. doi: 10.1007/BF00248887. [DOI] [PubMed] [Google Scholar]
- Weaver P. F., Wall J. D., Gest H. Characterization of Rhodopseudomonas capsulata. Arch Microbiol. 1975 Nov 7;105(3):207–216. doi: 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]
- Wu L. F., Mandrand M. A. Microbial hydrogenases: primary structure, classification, signatures and phylogeny. FEMS Microbiol Rev. 1993 Apr;10(3-4):243–269. doi: 10.1111/j.1574-6968.1993.tb05870.x. [DOI] [PubMed] [Google Scholar]
- Zürrer H., Bachofen R. Hydrogen Production by the Photosynthetic Bacterium Rhodospirillum rubrum. Appl Environ Microbiol. 1979 May;37(5):789–793. doi: 10.1128/aem.37.5.789-793.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zwaan J. W., Coremans J. M., Bouwens E. C., Albracht S. P. Effect of 17O2 and 13CO on EPR spectra of nickel in hydrogenase from Chromatium vinosum. Biochim Biophys Acta. 1990 Nov 15;1041(2):101–110. doi: 10.1016/0167-4838(90)90051-g. [DOI] [PubMed] [Google Scholar]



