Abstract
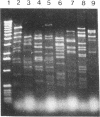
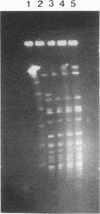
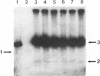
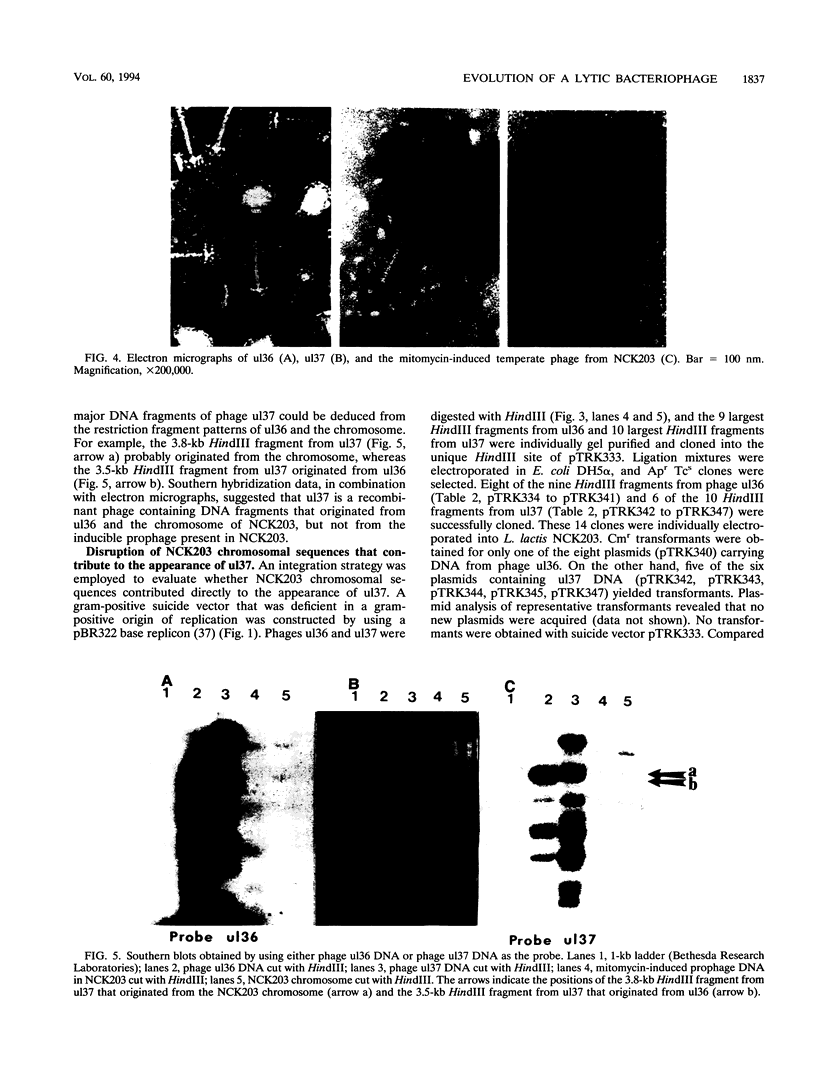
We discovered a phage-host interaction in which the lytic phage ul36, in response to pressure exerted by an abortive phage resistance mechanism, acquired a large DNA fragment from the chromosome of Lactococcus lactis NCK203 to form a new phage, ul37. Phage ul37 was characterized at morphological, phenotypic, and genotypic levels and was found to be a member of the P335 species. Although it exhibits a high level of DNA homology with ul36, phage ul37 is resistant to the abortive mechanism and has a longer tail, a different base plate, and apparently a different origin of replication. The chromosomal DNA implicated in the formation of new phage ul37 was disrupted by site-specific integration in NCK203. This strategy prevented the appearance of ul37 during subsequent infections with ul36.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alatossava T., Klaenhammer T. R. Molecular Characterization of Three Small Isometric-Headed Bacteriophages Which Vary in Their Sensitivity to the Lactococcal Phage Resistance Plasmid pTR2030. Appl Environ Microbiol. 1991 May;57(5):1346–1353. doi: 10.1128/aem.57.5.1346-1353.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J., Daly C., Fitzgerald G. F. Controlled Integration into the Lactococcus Chromosome of the pCI829-Encoded Abortive Infection Gene from Lactococcus lactis subsp. lactis UC811. Appl Environ Microbiol. 1992 Oct;58(10):3283–3291. doi: 10.1128/aem.58.10.3283-3291.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Chopin M. C., Chopin A., Rouault A., Galleron N. Insertion and amplification of foreign genes in the Lactococcus lactis subsp. lactis chromosome. Appl Environ Microbiol. 1989 Jul;55(7):1769–1774. doi: 10.1128/aem.55.7.1769-1774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin M. C. Resistance of 17 mesophilic lactic Streptococcus bacteriophages to pasteurization and spray-drying. J Dairy Res. 1980 Feb;47(1):131–139. doi: 10.1017/s0022029900020963. [DOI] [PubMed] [Google Scholar]
- Coveney J. A., Fitzgerald G. F., Daly C. Detailed characterization and comparison of four lactic streptococcal bacteriophages based on morphology, restriction mapping, DNA homology, and structural protein analysis. Appl Environ Microbiol. 1987 Jul;53(7):1439–1447. doi: 10.1128/aem.53.7.1439-1447.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B. E., Powell I. B., Hillier A. J. Temperate bacteriophages and lysogeny in lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):79–90. doi: 10.1111/j.1574-6968.1990.tb04880.x. [DOI] [PubMed] [Google Scholar]
- Duckworth D. H., Glenn J., McCorquodale D. J. Inhibition of bacteriophage replication by extrachromosomal genetic elements. Microbiol Rev. 1981 Mar;45(1):52–71. doi: 10.1128/mr.45.1.52-71.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmaz E., Higgins D. L., Klaenhammer T. R. Molecular characterization of a second abortive phage resistance gene present in Lactococcus lactis subsp. lactis ME2. J Bacteriol. 1992 Nov;174(22):7463–7469. doi: 10.1128/jb.174.22.7463-7469.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington A., Hill C. Construction of a Bacteriophage-Resistant Derivative of Lactococcus lactis subsp. lactis 425A by Using the Conjugal Plasmid pNP40. Appl Environ Microbiol. 1991 Dec;57(12):3405–3409. doi: 10.1128/aem.57.12.3405-3409.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. L., Sanozky-Dawes R. B., Klaenhammer T. R. Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J Bacteriol. 1988 Aug;170(8):3435–3442. doi: 10.1128/jb.170.8.3435-3442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Massey I. J., Klaenhammer T. R. Rapid method to characterize lactococcal bacteriophage genomes. Appl Environ Microbiol. 1991 Jan;57(1):283–288. doi: 10.1128/aem.57.1.283-288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J Bacteriol. 1990 Nov;172(11):6419–6426. doi: 10.1128/jb.172.11.6419-6426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J Bacteriol. 1991 Jul;173(14):4363–4370. doi: 10.1128/jb.173.14.4363-4370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Pierce K., Klaenhammer T. R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction modification (R+/M+) and abortive infection (Hsp+). Appl Environ Microbiol. 1989 Sep;55(9):2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. DNA-DNA Homology Between Lactic Streptococci and Their Temperate and Lytic Phages. Appl Environ Microbiol. 1984 May;47(5):1031–1038. doi: 10.1128/aem.47.5.1031-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Fitzgerald G. F., Mata M., Mercenier A., Neve H., Powell I. B., Ronda C., Saxelin M., Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32(1):2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- Jarvis A. W., Heap H. A., Limsowtin G. K. Resistance against Industrial Bacteriophages Conferred on Lactococci by Plasmid pAJ1106 and Related Plasmids. Appl Environ Microbiol. 1989 Jun;55(6):1537–1543. doi: 10.1128/aem.55.6.1537-1543.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Meyer J. Electron microscopic heteroduplex study and restriction endonuclease cleavage analysis of the DNA genomes of three lactic streptococcal bacteriophages. Appl Environ Microbiol. 1986 Mar;51(3):566–571. doi: 10.1128/aem.51.3.566-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. Serological studies of a host range mutant of a lactic streptococcal bacteriophage. Appl Environ Microbiol. 1978 Dec;36(6):785–789. doi: 10.1128/aem.36.6.785-789.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R. Development of bacteriophage-resistant strains of lactic acid bacteria. Biochem Soc Trans. 1991 Aug;19(3):675–681. doi: 10.1042/bst0190675. [DOI] [PubMed] [Google Scholar]
- Kok J., van der Vossen J. M., Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984 Oct;48(4):726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. H., Bickle T. A. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev. 1983 Sep;47(3):345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K. J., Gietema J., Kok J., Venema G. Chromosomal stabilization of the proteinase genes in Lactococcus lactis. Appl Environ Microbiol. 1991 Sep;57(9):2568–2575. doi: 10.1128/aem.57.9.2568-2575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moineau S., Bernier D., Jobin M., Hébert J., Klaenhammer T. R., Pandian S. Production of Monoclonal Antibodies against the Major Capsid Protein of the Lactococcus Bacteriophage ul36 and Development of an Enzyme-Linked Immunosorbent Assay for Direct Phage Detection in Whey and Milk. Appl Environ Microbiol. 1993 Jul;59(7):2034–2040. doi: 10.1128/aem.59.7.2034-2040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moineau S., Durmaz E., Pandian S., Klaenhammer T. R. Differentiation of Two Abortive Mechanisms by Using Monoclonal Antibodies Directed toward Lactococcal Bacteriophage Capsid Proteins. Appl Environ Microbiol. 1993 Jan;59(1):208–212. doi: 10.1128/aem.59.1.208-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moineau S., Pandian S., Klaenhammer T. R. Restriction/Modification systems and restriction endonucleases are more effective on lactococcal bacteriophages that have emerged recently in the dairy industry. Appl Environ Microbiol. 1993 Jan;59(1):197–202. doi: 10.1128/aem.59.1.197-202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'sullivan D. J., Hill C., Klaenhammer T. R. Effect of Increasing the Copy Number of Bacteriophage Origins of Replication, in trans, on Incoming-Phage Proliferation. Appl Environ Microbiol. 1993 Aug;59(8):2449–2456. doi: 10.1128/aem.59.8.2449-2456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'sullivan D. J., Klaenhammer T. R. Rapid Mini-Prep Isolation of High-Quality Plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol. 1993 Aug;59(8):2730–2733. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell I. B., Davidson B. E. Characterization of streptococcal bacteriophage c6A. J Gen Virol. 1985 Dec;66(Pt 12):2737–2741. doi: 10.1099/0022-1317-66-12-2737. [DOI] [PubMed] [Google Scholar]
- Powell I. B., Davidson B. E. Resistance to In Vitro Restriction of DNA from Lactic Streptococcal Bacteriophage c6A. Appl Environ Microbiol. 1986 Jun;51(6):1358–1360. doi: 10.1128/aem.51.6.1358-1360.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevots F., Mata M., Ritzenthaler P. Taxonomic differentiation of 101 lactococcal bacteriophages and characterization of bacteriophages with unusually large genomes. Appl Environ Microbiol. 1990 Jul;56(7):2180–2185. doi: 10.1128/aem.56.7.2180-2185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relano P., Mata M., Bonneau M., Ritzenthaler P. Molecular characterization and comparison of 38 virulent and temperate bacteriophages of Streptococcus lactis. J Gen Microbiol. 1987 Nov;133(11):3053–3063. doi: 10.1099/00221287-133-11-3053. [DOI] [PubMed] [Google Scholar]
- Reyrolle J., Chopin M. C., Letellier F., Novel G. Lysogenic strains of lactic Acid streptococci and lytic spectra of their temperate bacteriophages. Appl Environ Microbiol. 1982 Feb;43(2):349–356. doi: 10.1128/aem.43.2.349-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Restriction and modification in group N streptococci: effect of heat on development of modified lytic bacteriophage. Appl Environ Microbiol. 1980 Sep;40(3):500–506. doi: 10.1128/aem.40.3.500-506.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Leonhard P. J., Sing W. D., Klaenhammer T. R. Conjugal strategy for construction of fast Acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986 Nov;52(5):1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol. 1986 Jan;3(1):75–83. doi: 10.1093/oxfordjournals.molbev.a040377. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tanskanen E. I., Tulloch D. L., Hillier A. J., Davidson B. E. Pulsed-Field Gel Electrophoresis of SmaI Digests of Lactococcal Genomic DNA, a Novel Method of Strain Identification. Appl Environ Microbiol. 1990 Oct;56(10):3105–3111. doi: 10.1128/aem.56.10.3105-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap W. Y., Kreuzer K. N. Recombination hotspots in bacteriophage T4 are dependent on replication origins. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6043–6047. doi: 10.1073/pnas.88.14.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]