Abstract
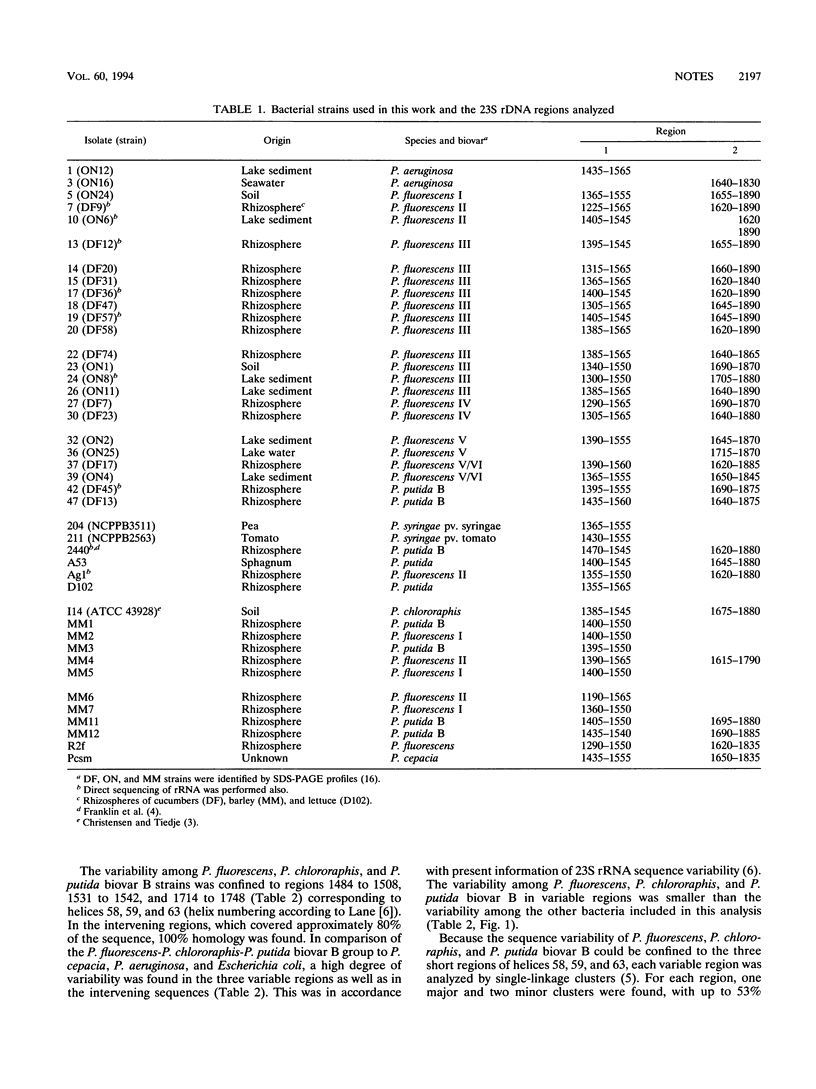
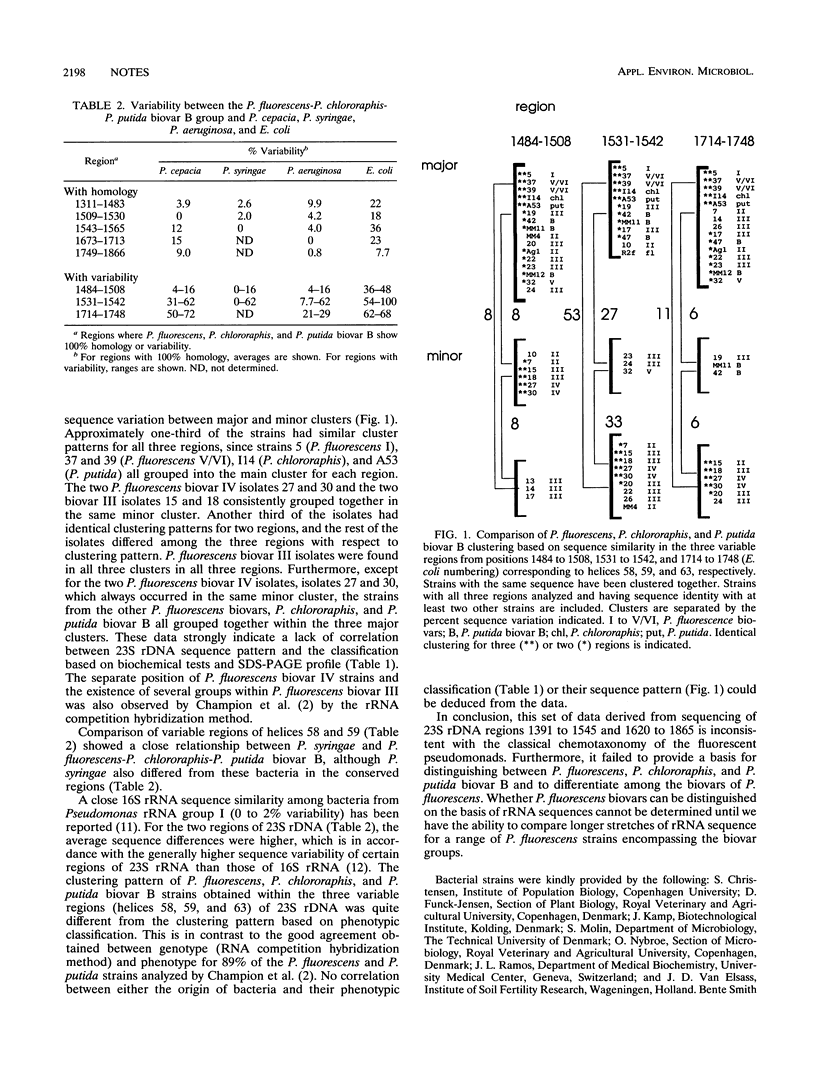
The regions from positions 1391 to 1545 and 1620 to 1865 (Escherichia coli numbers) of the 23S ribosomal DNA sequences have been analyzed for a number of Pseudomonas fluorescens and P. putida isolates. Variability was observed only in three smaller regions from positions 1484 to 1508, 1531 to 1542, and 1714 to 1748, corresponding to helices 58, 59, and 63, respectively, where up to 53% dissimilarity was found. Sequence analysis did not allow clear distinctions among P. fluorescens biovars, P. chlororaphis, and P. putida biovar B.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett E. L., Solanes R. E., Tang J. S., Palleroni N. J. Pseudomonas fluorescens biovar V: its resolution into distinct component groups and the relationship of these groups to other P. fluorescens biovars, to P. putida, and to psychrotrophic pseudomonads associated with food spoilage. J Gen Microbiol. 1986 Oct;132(10):2709–2721. doi: 10.1099/00221287-132-10-2709. [DOI] [PubMed] [Google Scholar]
- Champion A. B., Barrett E. L., Palleroni N. J., Soderberg K. L., Kunisawa R., Contopoulou R., Wilson A. C., Doudoroff M. Evolution in Pseudomonas fluorescens. J Gen Microbiol. 1980 Oct;120(2):485–511. doi: 10.1099/00221287-120-2-485. [DOI] [PubMed] [Google Scholar]
- Christensen S., Tiedje J. M. Sub-Parts-Per-Billion Nitrate Method: Use of an N(2)O-Producing Denitrifier to Convert NO(3) or NO(3) to N(2)O. Appl Environ Microbiol. 1988 Jun;54(6):1409–1413. doi: 10.1128/aem.54.6.1409-1413.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J. Phylogenetic analysis using ribosomal RNA. Methods Enzymol. 1988;164:793–812. doi: 10.1016/s0076-6879(88)64084-5. [DOI] [PubMed] [Google Scholar]
- Rasmussen O. F., Beck T., Olsen J. E., Dons L., Rossen L. Listeria monocytogenes isolates can be classified into two major types according to the sequence of the listeriolysin gene. Infect Immun. 1991 Nov;59(11):3945–3951. doi: 10.1128/iai.59.11.3945-3951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands D. C., Rovira A. D. Pseudomonas fluorescens biotype G, the dominant fluorescent pseudomonad in South Australian soils and wheat rhizospheres. J Appl Bacteriol. 1971 Mar;34(1):261–275. doi: 10.1111/j.1365-2672.1971.tb02285.x. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


