Abstract
The presence of Frankia strains in soil samples collected from northern areas of Pakistan was detected by inoculating Coriaria nepalensis and Datisca cannabina plants. The abundance of compatible Frankia strains in some areas was indicated by profuse nodulation of the host plants, whereas soil samples from other localities failed to result in nodulation. An oligonucleotide probe (COR/DAT) directed against the 16S rRNA gene of the endophytes of Coriaria and Datisca spp. that did not cross-react with the RNA gene of Frankia strains isolated from other hosts was developed. Genetic diversity among Frankia strains nodulating D. cannabina was determined by sequence analysis of the partial 16S rRNA gene amplified from nodules induced by soil samples from different localities by PCR. Four types of Frankia sequences and one non-Frankia sequence were detected by hybridization with a Frankia genus probe and the COR/DAT probe as well as by sequence analysis of the cloned PCR products.
Full text
PDF

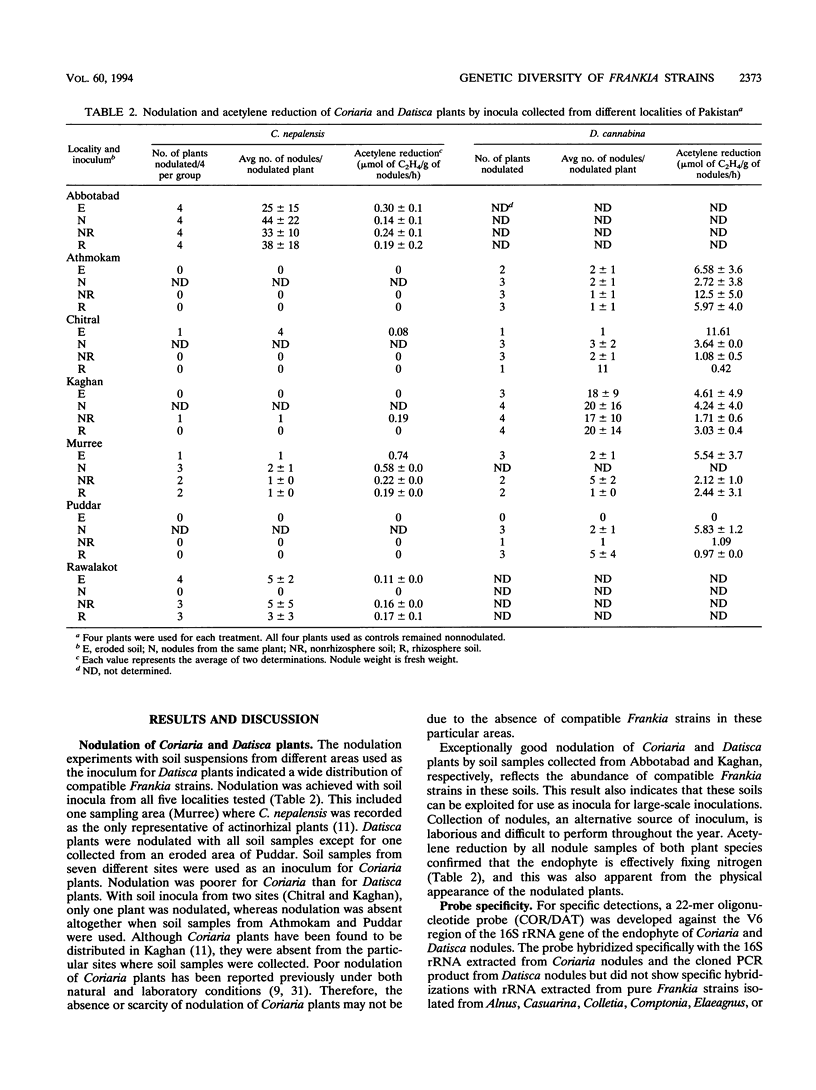
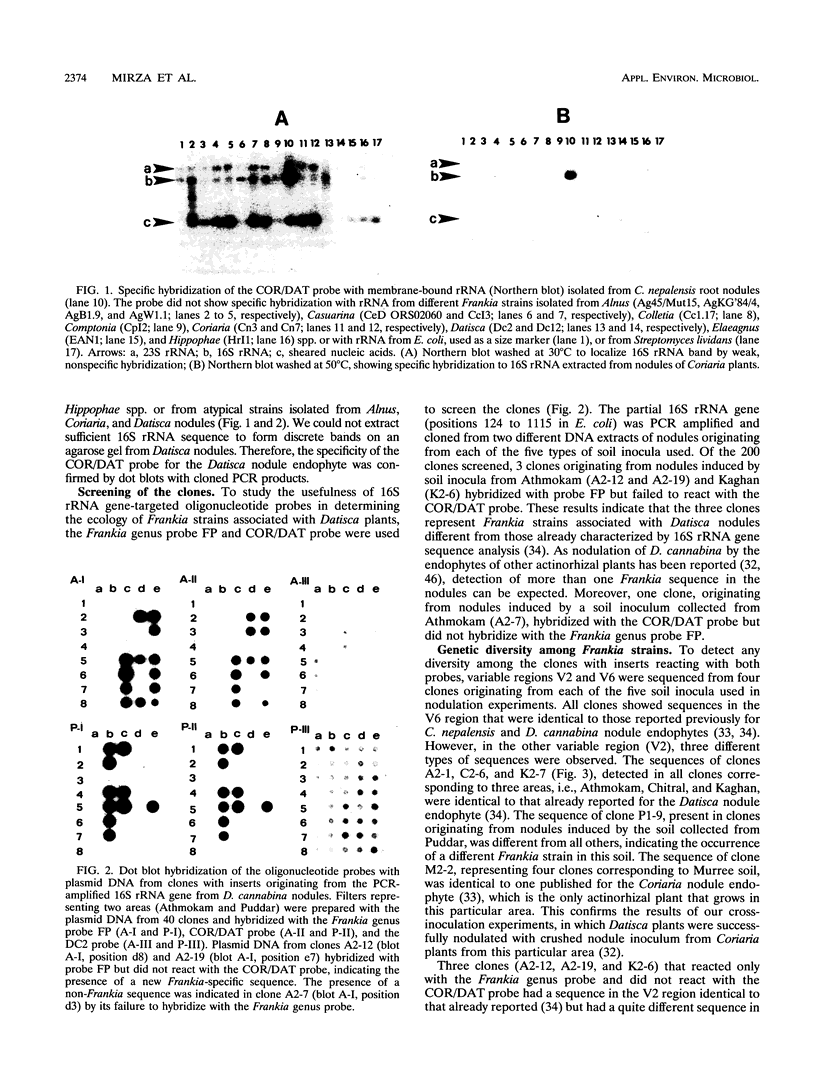


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benson D. R., Buchholz S. E., Hanna D. G. Identification of frankia strains by two-dimensional polyacrylamide gel electrophoresis. Appl Environ Microbiol. 1984 Mar;47(3):489–494. doi: 10.1128/aem.47.3.489-494.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom R. A., Mullin B. C., Tate R. L., 3rd DNA restriction patterns and DNA-DNA solution hybridization studies of Frankia isolates from Myrica pennsylvanica (bayberry). Appl Environ Microbiol. 1989 Sep;55(9):2155–2160. doi: 10.1128/aem.55.9.2155-2160.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaham D., Deltredici P., Torrey J. G. Isolation and Cultivation in vitro of the Actinomycete Causing Root Nodulation in Comptonia. Science. 1978 Feb 24;199(4331):899–902. doi: 10.1126/science.199.4331.899. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe J. S., Cooper H. J., Smith M. A., Sims M. J., Parker D., Gewert D. Improved cloning efficiency of polymerase chain reaction (PCR) products after proteinase K digestion. Nucleic Acids Res. 1991 Jan 11;19(1):184–184. doi: 10.1093/nar/19.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley T. M., Smida J., Stackebrandt E. Reverse transcriptase sequencing of 16S ribosomal RNA from Faenia rectivirgula, Pseudonocardia thermophila and Saccharopolyspora hirsuta, three wall type IV actinomycetes which lack mycolic acids. J Gen Microbiol. 1988 Apr;134(4):961–966. doi: 10.1099/00221287-134-4-961. [DOI] [PubMed] [Google Scholar]
- Giovannoni S. J., Britschgi T. B., Moyer C. L., Field K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990 May 3;345(6270):60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Hahn D., Kester R., Starrenburg M. J., Akkermans A. D. Extraction of ribosomal RNA from soil for detection of Frankia with oligonucleotide probes. Arch Microbiol. 1990;154(4):329–335. doi: 10.1007/BF00276527. [DOI] [PubMed] [Google Scholar]
- Nazaret S., Cournoyer B., Normand P., Simonet P. Phylogenetic relationships among Frankia genomic species determined by use of amplified 16S rDNA sequences. J Bacteriol. 1991 Jul;173(13):4072–4078. doi: 10.1128/jb.173.13.4072-4078.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., De Rijk P., Goris A., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):1987–2015. doi: 10.1093/nar/19.suppl.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick G., Paget E., Simonet P., Moiroud A., Normand P. The nodular endophytes of Coriaria spp. form a distinct lineage within the genus Frankia. Mol Ecol. 1992 Oct;1(3):175–181. doi: 10.1111/j.1365-294x.1992.tb00173.x. [DOI] [PubMed] [Google Scholar]
- Normand P., Cournoyer B., Simonet P., Nazaret S. Analysis of a ribosomal RNA operon in the actinomycete Frankia. Gene. 1992 Feb 1;111(1):119–124. doi: 10.1016/0378-1119(92)90612-s. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Tohdoh N., Sugiura M. The complete nucleotide sequence of 16S ribosomal RNA gene from tobacco chloroplasts. Gene. 1982 Feb;17(2):213–218. doi: 10.1016/0378-1119(82)90074-9. [DOI] [PubMed] [Google Scholar]
- Ward D. M., Weller R., Bateson M. M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990 May 3;345(6270):63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro F. B., Miguel C., Rodriguez-Barrueco C. A study of the capacity of soils to induce nodules in Alnus glutinosa (L.) Gaertn. and Myrica gale L., with special reference to the specificity of the endophytes. Ann Microbiol (Paris) 1976 Feb-Mar;127A(2):307–315. [PubMed] [Google Scholar]