Abstract
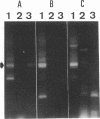
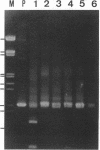
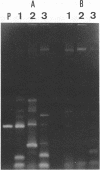
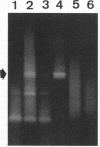
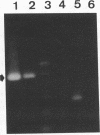
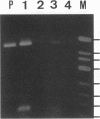
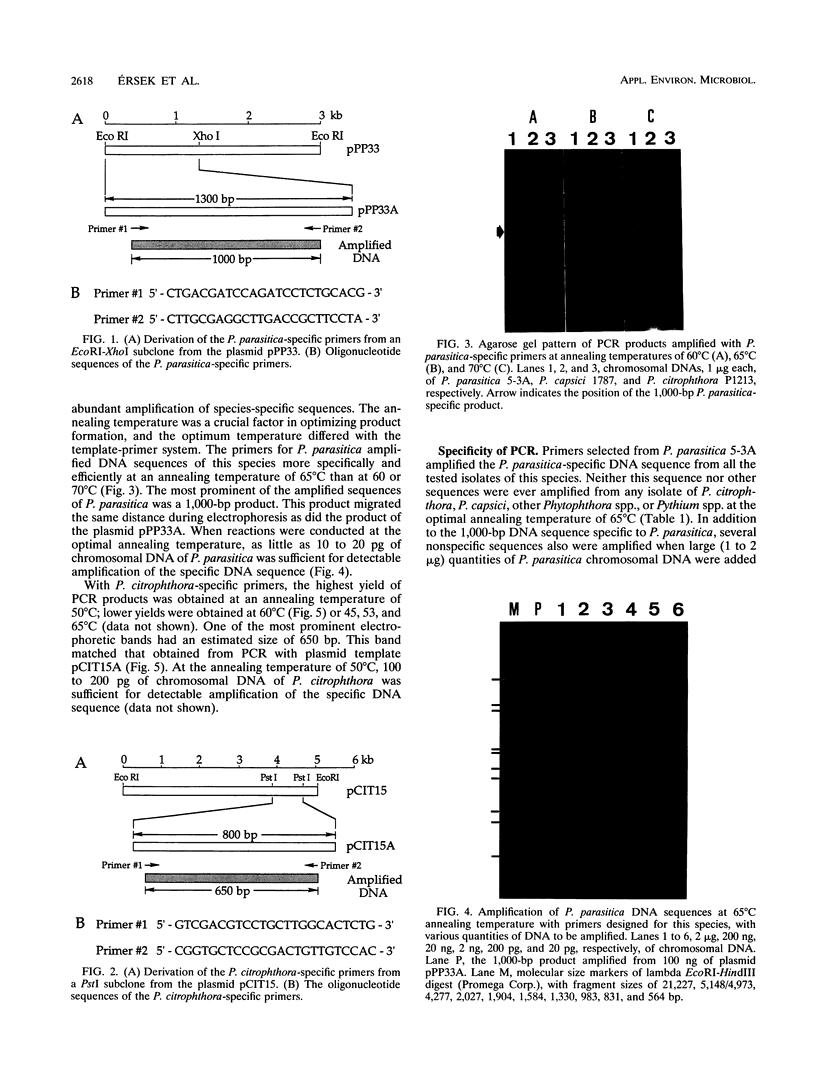
We used PCR to differentiate species in the genus Phytophthora, which contains a group of devastating plant pathogenic fungi. We focused on Phytophthora parasitica, a species that can infect solanaceous plants such as tomato, and on Phytophthora citrophthora, which is primarily a citrus pathogen. Oligonucleotide primers were derived from sequences of a 1,300-bp P. parasitica-specific DNA segment and of an 800-bp P. citrophthora-specific segment. Under optimal conditions, the primers developed for P. parasitica specifically amplified a 1,000-bp sequence of DNA from isolates of P. parasitica. Primers for P. citrophthora similarly and specifically amplified a 650-bp sequence of DNA from isolates of P. citrophthora. Detectable amplification of these specific DNA sequences required picogram quantities of chromosomal DNA. Neither pair of primers amplified these sequences with DNAs from other species of Phytophthora or from the related genus Pythium. DNAs from P. parasitica and P. citrophthora growing in infected tomato stem tissue were amplified as distinctly as DNAs from axenic cultures of each fungal species. This is the first report on PCR-driven amplification with Phytophthora species-specific primers.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailey A. M., Mena G. L., Herrera-Estrella L. Genetic transformation of the plant pathogens Phytophthora capsici and Phytophthora parasitica. Nucleic Acids Res. 1991 Aug 11;19(15):4273–4278. doi: 10.1093/nar/19.15.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin P. H., Kirkpatrick B. C., Duniway J. M. Identification of Phytophthora citrophthora with Cloned DNA Probes. Appl Environ Microbiol. 1990 Mar;56(3):669–674. doi: 10.1128/aem.56.3.669-674.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson H. S., Tyler B. M., Michelmore R. W. Transformation of the oomycete pathogen, Phytophthora infestans. Mol Plant Microbe Interact. 1991 Nov-Dec;4(6):602–607. doi: 10.1094/mpmi-4-602. [DOI] [PubMed] [Google Scholar]
- Rollo F., Salvi R., Torchia P. Highly sensitive and fast detection of Phoma tracheiphila by polymerase chain reaction. Appl Microbiol Biotechnol. 1990 Feb;32(5):572–576. doi: 10.1007/BF00173730. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schesser K., Luder A., Henson J. M. Use of Polymerase Chain Reaction To Detect the Take-All Fungus, Gaeumannomyces graminis, in Infected Wheat Plants. Appl Environ Microbiol. 1991 Feb;57(2):553–556. doi: 10.1128/aem.57.2.553-556.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]