Abstract
The x-ray structures of an inhibitor complex of the catalytic core domain of avian sarcoma virus integrase (ASV IN) were solved at 1.9- to 2.0-Å resolution at two pH values, with and without Mn2+ cations. This inhibitor (Y-3), originally identified in a screen for inhibitors of the catalytic activity of HIV type 1 integrase (HIV-1 IN), was found in the present study to be active against ASV IN as well as HIV-1 IN. The Y-3 molecule is located in close proximity to the enzyme active site, interacts with the flexible loop, alters loop conformation, and affects the conformations of active site residues. As crystallized, a Y-3 molecule stacks against its symmetry-related mate. Preincubation of IN with metal cations does not prevent inhibition, and Y-3 binding does not prevent binding of divalent cations to IN. Three compounds chemically related to Y-3 also were investigated, but no binding was observed in the crystals. Our results identify the structural elements of the inhibitor that likely determine its binding properties.
Retroviral integrase (IN)‖ catalyzes the integration of reverse-transcribed viral DNA into the host genome in two steps called processing and joining (1–3). These reactions are chemically similar, proceeding via nucleophilic attack on the DNA by a donor hydroxyl group (water or ribose 3′ OH), activated by the catalytic site of the enzyme. In vitro, these reactions require only divalent cations and water as cofactors. The in vitro reactions are fastest with Mn2+ as the metal cofactor, although Mg2+ also can be used. Because of its greater abundance in living cells, Mg2+ is assumed to be the cofactor in vivo. Previous structural studies using the catalytic core domain of avian sarcoma virus (ASV) IN showed the presence of one cation of Mn2+ or Mg2+, or two cations of Zn2+ in the active site (4, 5).
Retroviral INs are ≈300 amino acids in length and comprised three domains. The N-terminal domain (residues 1 to ≈50) binds and is stabilized by Zn2+ (6–8). The C-terminal domain (residues ≈200–300) binds DNA in a sequence-independent fashion (9). The catalytic core domain (residues ≈50–200) contains the active site triad consisting of residues Asp-64, Asp-121, and Glu-157. This triad is essential for enzymatic activity and binds the required metal cofactors (4, 5). The structures of the C- and N-terminal domains of HIV-1 IN were solved by nuclear magnetic resonance (10–12), whereas the core domains of HIV IN and ASV IN were determined by x-ray crystallography (13, 14). Currently, no structural information is available for multi-domain fragments of IN.
Through its role in integration, IN is essential for the retroviral life cycle. Thus, IN is well suited as a target for drug design, especially as no analogous host enzymes are known (15). No structural data on the binding of inhibitors, or even divalent cations, to HIV-1 IN have been published. For this reason, ASV IN has become an important target for structural studies. Here, we present the first structure of an inhibitor, Y-3, complexed to the catalytic core of ASV IN. Crystallization of the ASV IN–Y-3 complex thus has implications for directed antiviral drug design.
MATERIALS AND METHODS
Preparation and Analysis of Inhibitors.
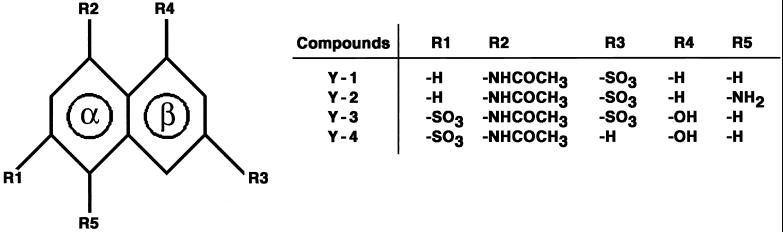
To identify new HIV-1 IN inhibitors, we tested many compounds that showed activity in the National Cancer Institute’s (NCI) antiviral drug screen against CEM-infected cells (virally infected cell line designed for antiviral screening assays). Several aromatic sulfones, disulfides, sulfonates, and sulfonamides were shown to inhibit IN at low micromolar concentrations (reviewed in ref. 16). Starting from known inhibitors including caffeic acid phenethyl ester and an anthraquinone derivative (NSC115290) (17), we derived a putative three-point pharmacophore model that was used to screen the National Cancer Institute three-dimensional database of ≈200,000 compounds (18). Compounds Y-1, Y-2, and Y-3 were identified in that search. Compounds Y-1, 5-(acetylamino)naphthalene-2-sulfonic acid; Y-2, 5-acetylamino-8-aminonaphthalene-2-sulfonic acid; and Y-3, 4-acetylamino-5-hydroxynaphthalene-2,7-disulfonic acid were obtained from the NCI Chemical Repository through the Drug Synthesis and Chemistry Branch. Compound Y-4, 4-acetylamino-5-hydroxynaphthalene-2-sulfonic acid (monosodium salt) (Mr = 281) was synthesized for this study by conversion of 8-aminonaphthalene-1,6-disulfonic acid to 4-amino-5-hydroxynaphthalene-2-disulfonic (sodium salt), using a published procedure (19). The product was acetylated using acetic anhydride in anhydrous pyridine. All of the four compounds used for these experiments are derivatives of naphthalene (Fig. 1). For simplicity, alpha (α) refers to the aromatic ring with substituents R1, R2, and R5, and beta (β) refers to the ring with substituents R3 and R4.
Figure 1.
Inhibitor compounds. A general formula is shown at left. The aromatic rings of the naphthalene system are marked as α and β. Definitions of groups R1–R5 are shown at right.
HIV-1 and ASV IN Assays.
The 3′-processing and 3′-end-joining reactions of HIV-1 IN were assayed as described (20). IC50 values were determined by plotting inhibitor concentration vs. percent inhibition and calculating the concentration that produced 50% inhibition. Disintegration reactions were performed as described above with an oligonucleotide (i.e., branched substrate) in which the U5 end was “integrated” into target DNA (21).
The 3′-processing activity of full-length ASV IN (1–286) and the endonucleolytic activity of the isolated catalytic core of ASV IN (52–207) were assayed as described (22) using the U3 end of linear ASV DNA [U3(+), 5′-AAT GTA GTC TTA TGC AAT-3′; U3(-), 5′-ATT GCA TAA GAC TAC ATT-3′]. ASV IN (1–286) was tested with 10 mM MnCl2, 10 mM MgCl2, or 1 mM ZnCl2; ASV IN (52–207) was tested with 10 mM MnCl2 or 2 mM ZnCl2, optimal for the respective enzymatic activities (5). Final concentrations of other components in each reaction (10 μl) were 2 μM ASV IN proteins, 15 μM 32P-labeled oligonucleotide substrate, 50 mM Hepes (pH 8.2), 2 mM 2-mercaptoethanol, 0.2 mg/ml BSA, 5 mM Hepes (pH 8.1), 50 mM NaCl, 0.1% thiodiglycol, 0.01 mM EDTA, and 4% glycerol. For inhibition studies, each prospective inhibitor was incubated with enzyme and divalent cation cofactor for 30 min at 30°C, double-stranded oligonucleotide substrate was added, and incubation continued at 37°C (see Fig. 2 legend for details). Reactions were stopped by the addition of 10 μl of 25 mM EDTA, dried, and analyzed by electrophoresis in a denaturing 20% acrylamide gel.
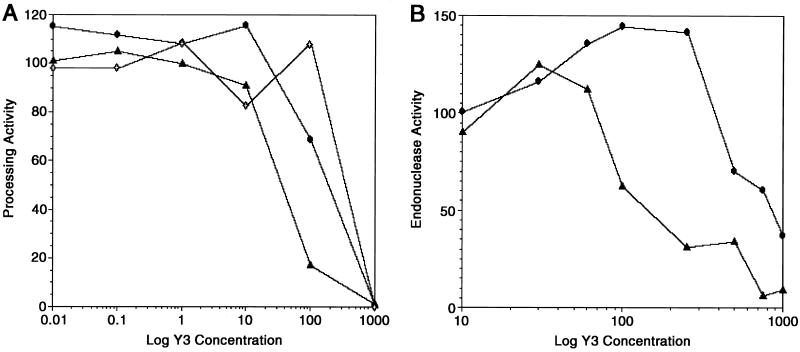
Figure 2.
Inhibition of processing and endonuclease activities of ASV IN proteins by Y-3. (A) Processing activity of ASV IN (1–286) measured using Mn2+ (circles), Mg2+ (open diamonds), or Zn2+(triangles) as cofactor, with increasing concentrations of Y-3. Reactions containing Mn2+ were incubated at 37°C for 10 min.; those containing either Mg2+ or Zn2+ were incubated for 60 min. Processing activity is expressed as relative percentage of positive (no inhibitor) control. (B) Endonucleolytic activity of ASV IN (52–207) was measured using Mn2+(circles) or Zn2+(triangles), with increasing concentrations of Y-3. Reactions containing Mn2+ were incubated as above; those containing Zn2+ were incubated for 120 min. Data are expressed as in A.
Crystallization, Soaking Experiments, Data Collection, and Structure Analysis.
ASV IN core domain preparation and crystallization have been described (14). Crystals of average linear dimensions 0.4–0.7 mm were grown in citrate buffer, pH 5.6. Potential inhibitors were dissolved in mother liquor up to a concentration of 0.1 M with either citrate (pH 5.6) or Hepes (pH 7.5) buffer. Higher concentrations of Y-2 and Y-3 (0.1–0.3 M) also were used with identical results. Some crystals were transferred to Y-3 inhibitor (0.3 M), Hepes (pH 7.5), and MnCl2 (0.025–0.15 M). All crystallization (≈1 week) and soaking (≈2–3 weeks) experiments were performed at 5°C.
X-ray data were collected at room temperature for crystals soaked in each of four compounds (Y-1, Y-2, Y-3[±MnCl2], and Y-4), in both the citrate and Hepes buffers. Experiments were repeated at least twice to control for any variations in inhibitor concentration during soaking. Diffraction data were measured using CuKα radiation from a Rigaku (Danvers, MA) RU200 rotating anode generator and recorded on a DIP2020 image plate detector (Nonius, Delft, The Netherlands). For most data sets, the high resolution diffraction limit extended to ≈2.0 Å with good scaling statistics. All of the data sets were processed and scaled with the HKL suite of programs (23); see Table 1 for statistics.
Table 1.
Data collection and refinement statistics for ASV IN core domain complex with inhibitor Y-3
| Measurements | pH 5.6 (citrate)
|
pH 7.5 Hepes
|
|
|---|---|---|---|
| No metal bound | No metal bound | Mn2+ bound | |
| Unit cell dimensions (a, c) Å | 66.71, 81.02 | 66.44, 81.28 | 66.44, 81.07 |
| Highest resolution of x-ray data, Å | 2.0 | 1.9 | 1.9 |
| Total number of reflections | 93168 | 115637 | 92081 |
| Unique reflections, n | 12682 | 14942 | 14653 |
| Completeness, % | 99.0 | 99.9 | 98.2 |
| Rsym*, % | 7.2 | 5.8 | 6.6 |
| Rcryst†, % | 15.1 | 15.5 | 15.2 |
| Rfree†, % | 22.2 | 21.2 | 21.0 |
| Rms deviation from ideal geometry | |||
| Bond lengths, Å | 0.011 | 0.012 | 0.012 |
| Bond angles, ° | 1.42 | 1.67 | 1.62 |
| Improper angles, ° | 1.98 | 1.99 | 1.83 |
| Dihedral angles, ° | 23.31 | 23.37 | 23.30 |
| Average B-factor, Å2 | |||
| All nonhydrogen atoms | 30.85 | 30.26 | 29.65 |
| Inhibitor atoms | 31.33 | 42.17 | 31.54 |
| Nonhydrogen atoms, n | |||
| All atoms | 1279 | 1316 | 1320 |
| Water molecules | 126 | 169 | 172 |
Rsym(I) = ΣΣ/I(I)−〈I(hkl)〉//Σ(I), summed over all reflections and observations, where I(I) is the ith observation of the hkl reflection intensity and 〈I(hkl)〉 is the mean hkl reflection intensity.
Rcryst(F) = Σh//Fobs(h)/−/Fcalc(h)///Σh/Fobs(h)/, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes for the reflection with Miller indices h = (h, k, l). The free R-factor (Rfree) is calculated for a random set of reflections (10%) excluded from the refinement.
In all cases, for refinement and electron density (ED) map calculations, x-ray data within the resolution range 8.0 Å to the highest available [F > 2.0⋅σ(F)] were used. A subset of data (≈10% of all reflections) was excluded from refinement and used for cross-validation with free R-factor (24). One structure of ASV IN, Protein Data Bank code 1ASV, was used always as the starting model after solvent was removed. Rigid body refinement was done to compensate for small variations in unit cell parameters. Difference Fourier maps (Fo–Fc and 2Fo–Fc) were calculated, and the structure was adjusted manually. Positional refinement, followed by refinement of isotropic B-factors for nonhydrogen atoms, was done for each model. Further refinement was done only in cases for which the difference ED maps indicated changes in the conformations of protein elements, the presence of inhibitor bound to IN, and/or bound Mn2+ cation. Amino acid side chains were adjusted to agree with the corresponding ED, solvent molecules were placed in sites distant from the inhibitor binding site, and metal cation included as appropriate. New ED maps were calculated after more refinement, providing clearer information about inhibitor binding (Fig. 3). The inhibitor molecule and missing solvent molecules were modeled into the ED map followed by additional refinement. All refinement and ED map calculations were performed with the program x-plor (25); visual inspection and manual modifications used molecular graphics packages o (26) and chain (27).
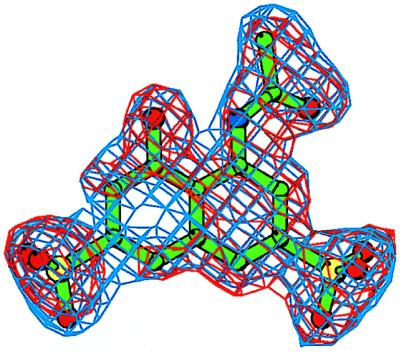
Figure 3.
An electron density (ED) map for the inhibitor Y-3. The initial Fo–Fc ED map is contoured at the 2.5σ level (red); the final 2Fo-Fc ED map is contoured at the 1.1σ level (blue). The refined conformation of the Y-3 model is shown in green. Prepared with bobscript, a modification of molscript (36).
RESULTS
Inhibition of HIV IN.
When tested in an inhibition assay specific for HIV-1 IN, the disulfonate Y-3 showed IC50 values of 16.2 μM for 3′-processing and 10.9 μM for strand transfer. In a disintegration assay, Y-3 also inhibited full-length HIV-1 IN with an IC50 value of ≈50 μM. Inhibition constants for the other three compounds were higher. Y-4 showed negligible inhibitory activity.
Inhibition of ASV IN.
Because we have shown previously that full-length ASV IN (1–286) and its isolated catalytic core [ASV IN (52–207)] are active with Zn2+ as a metal cofactor (5), we tested the inhibitor with this divalent cation in addition to Mn2+ or Mg2+. In all cases, Y-3 inhibited the processing activity of full-length protein in a concentration-dependent manner (Fig. 2A). IC50 values (≈40–350 μM) were consistent with those obtained with HIV-1 IN. Y-3 also inhibited endonuclease activity of the isolated catalytic core (52–207) (IC50 ≈120 μM with Zn2+ and 800 μM with Mn2+, Fig. 2B). There is no ASV IN catalytic core activity with Mg2+. A reproducible increase of catalytic activity was observed at Y-3 concentrations slightly lower than those that were inhibitory. The reason for this unexpected enhancement is unknown. In a separate experiment (not shown), we preincubated full-length and core proteins with either the inhibitor or the metal cofactor. No difference in the inhibitory activity was observed, suggesting that Y-3 and cations do not compete for the metal binding site.
Description of the Inhibitor Binding Site.
Of the four compounds investigated in this study, only Y-3 could be detected unambiguously in the ED map (Fig. 3). Based on the similarity of average temperature factors of the protein and inhibitor (Table 1), the binding site appears to be highly occupied. This site, located close to the crystallographic twofold axis, is formed by several residues of ASV IN located in the direct vicinity of the active site but not part of it (Fig. 4). Two molecules of Y-3 are observed close to each other, related by crystal symmetry, with an almost parallel arrangement of the naphthalene rings (≈15° deviation of their planes), and with distance between the centers of ≈4.4 Å. Each Y-3 molecule interacts with two crystallographically related monomers of ASV IN. It is convenient to categorize inhibitor contacts: Y-3 with one monomer of ASV IN; Y-3 with a second IN monomer; and Y-3–Y-3 (symmetry-related) inhibitor molecule.
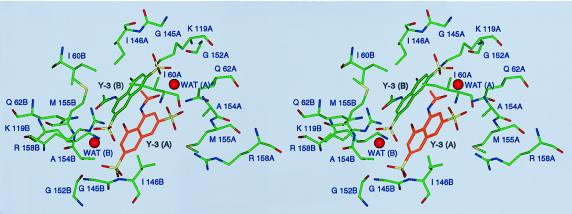
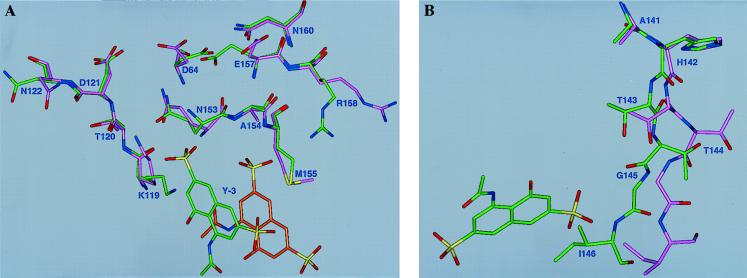
Figure 4.
Stereoview of the Y-3 binding site, shown approximately along the crystallographic twofold axis. Structural elements contributed by either monomer of the enzyme are labeled with A or B, respectively, after the residue number. Water molecules stabilizing the interactions between the two Y-3 molecules, WAT (A) and WAT (B), are shown.
The contacts between the R1 and R2 groups of Y-3 and the first IN monomer involve two oxygens of R1 within hydrogen bond distance of Arg-158 NH2 (3.1 Å), Gln-62 NE2 (2.8 Å), Ala-154 N (3.3 Å), and Met-155 N (3.1 Å). The Arg-158 hydrogen bond is rather weak, as deduced from its length and the existence of two alternate side chain orientations. The carbonyl oxygen of R2 forms hydrogen bonds with Lys-119 NZ (3.4 Å) and Glu-62 NE2 (2.9 Å). The methyl group of R2 is located in a hydrophobic pocket formed by the side chain of Ile-60 and the methyl group from the symmetry-related Y-3 molecule.
The symmetry-related second IN monomer interacts primarily with the R3 and R4 groups of the inhibitor. These interactions are weaker comparatively and stereochemically less well defined. The R3 group is in close contact with the main chain atoms of residues Ile-146, Gln-151, and Gly-152; only the first, between the R3 oxygen and Ile-146 N, is close enough (2.9 Å) to contribute significantly to Y-3 binding. The hydroxyl group R4 interacts with Lys-119 NZ (2.9 Å) and with a structural water molecule stabilized by interactions with protein atoms.
The third category of interactions consists of Y-3–Y-3 contacts. The α ring of one and β ring of the other molecule are involved in extensive π-like interactions, although the distance between the two rings (4.4 Å) is larger than typical for π-stacking [≈3.5 Å (28)] and the planes of interacting rings are not quite parallel. Stacking is probably stabilized by other hydrophobic interactions. A water molecule lying between the R1 sulfate and R2 of one Y-3 molecule and the R3 sulfate of the other symmetry-related Y-3 seems to add stability through hydrogen bonding, decreasing the energy strain from electrostatic interactions. Strong interactions between Y-3 and the first IN monomer (compared with the weaker interactions between Y-3 and the second IN monomer or second Y-3 molecule) suggests that in solution one monomer of IN binds a single Y-3 molecule. Additional ASV IN crystal forms or constructs will be needed to confirm this hypothesis.
Structural Changes in the Protein that Result from Inhibitor Binding.
The structure of the ASV IN core domain Y-3 inhibitor complex was compared with the previously determined ASV IN structure obtained under similar conditions [(4) - Protein Data Bank code 1ASV]. Several changes were observed, especially at lower pH. The most striking difference between the two structures at low pH is the orientation of the side chain of Asp-64, the central active site residue. In the apoenzyme, the side chain of this residue is oriented toward the side chain of Asp-121; in the inhibitor complex, it points in the opposite direction and forms two hydrogen bonds with the side chain of Asn-160 (Fig. 5A). In the absence of inhibitor, the interaction between the side chains of Asp-64 and Asp-121 is mediated by a water molecule. In the complex, a water molecule remains with Asp-121; however, an additional water molecule lies near the original position of Asp-64 OD1. At pH 7.5, the orientation of this side chain is identical in the complex and the native enzyme. The pH dependence of Asp-64 conformation is probably a direct consequence of changes in its protonation state.
Figure 5.
Inhibitor binding site. Conformational differences between the Y-3- ASV IN (core) complex [pH 5.6 (green)] and the unliganded enzyme, Protein Data Bank code 1ASV (magenta). (A) Residues with altered conformation are labeled. For Arg 158, one of two alternate conformations is shown. The bound Y-3 molecule and its symmetry-related copy are both shown. (B) Conformational changes in the flexible loop sections (residues 141–146) with bound inhibitor molecules.
Lys-119 also is affected by inhibitor binding (Fig. 5A). In the apoenzyme, its side chain is directed toward the center of the enzyme, forming a hydrogen bond with Thr-120 O, with a high B-factor indicating extensive side chain mobility. In the complex, this side chain reorients, turning away from the enzyme’s center and forming a direct hydrogen bond with the inhibitor. The B-factor for this side chain is ≈2 times lower in the complex than in the reference structure.
Residues 144–154 of ASV IN, highly conserved in different INs, form a flexible loop believed to be important in substrate binding (29–31). In 1ASV, most of the loop residues have very high B-factor values; in the ASV IN Y-3 complex, we were unable to determine loop structure for residues 147–152. In modeling this region (for completeness only), atomic occupancies were assigned zero values. Loop residues 144–146, however, showed clear conformational differences between the complex and 1ASV (Fig. 5B) with these three residues shifted toward the inhibitor molecule by up to 2.8 Å (for Cα of Ile-146). These changes also alter local solvent structure.
Another affected residue is Asn-122, which is separated from the inhibitor binding site by the flexible loop. The change in its conformation may be a direct consequence of shifting the flexible loop. The same may be true for Glu-157, whose side chain has an altered conformation and also interacts with the flexible loop. In the complex, Glu-157 OE2 points toward Asp-64 (≈3.7 Å), whereas Glu-157 OE1 is in a position occupied by a water molecule in the apoenzyme. We identified two conformations of the Arg-158 side chain. One conformation resembles Arg-158 in the apoenzyme, whereas in the other it approaches and hydrogen bonds with the inhibitor molecule. Finally, the binding of Y-3 alters the solvent structure in the inhibitor binding pocket, with two water molecules in the 1ASV structure substituted by the R1 and R2 groups of Y-3. The apoenzyme and complex are otherwise very similar, with minor structural differences limited to residues disordered in both structures.
Influence on Metal Binding.
To investigate possible interference between inhibitor and metal binding, we soaked ASV IN crystals in a solution containing Y-3 and Mn2+, each at 0.1 M. Because citrate anions form complexes with Mn2+, only Hepes (pH 7.5) was used. This pH change also may change residue conformations. Because of possible effects resulting from pH change, we soaked ASV IN crystals in Hepes buffer and Y-3, without Mn2+, as a control. The position and occupancy of the Mn2+-binding site were identical to the previously described structure (4). Inhibitor binding was similarly unaffected by pH change and Mn2+ binding, indicating both metal cation and inhibitor can bind simultaneously. This observation is consistent with our biochemical results, which showed that the order of metal cation and inhibitor addition does not affect inhibition. Additionally, residue conformations at pH 7.5 were the same with or without Mn2+.
This experiment allowed us to look at the effects of pH/buffer in the absence of inhibitor. Several residues have different side chain conformations at pH 7.5 (Hepes buffer) vs. pH 5.6 (citrate buffer). Besides the previously noted conformational change of the Asp-64 side chain, Arg-158 and Asn-122 also showed altered side chain conformations. At pH 7.5, Arg-158 appears in a single conformation, similar to that in the apoenzyme. The conformation of Asn-122 also was different, but this residue also shows a significant increase of B-factor values at the higher pH.
Residues 144–146 and Lys-119 were unaffected by pH or metal binding, but in the presence of the inhibitor, their conformations were different than in the apoenzyme. This result suggests that the binding of Y-3 alters the conformation of the flexible loop and of residues interacting with this loop. It is possible Y-3 inhibits ASV IN by interfering with the dynamic properties of this highly conserved loop and thus disrupting the interaction of the loop with the substrate molecule (31).
Mode of Binding and Structural Comparisons.
Initially, our studies included only three compounds, Y-1 through Y-3, identified as potential inhibitors of HIV-1 IN during a search of the NCI 3D database. We extended the study to Y-4 (related to Y-3 but lacking the R3 sulfonate), which we synthesized to examine the importance of these groups in Y-3 binding and inhibition. The four compounds are chemically similar, so why is only Y-3 observed to bind to the enzyme? Only Y-3 contains two sulfonate groups (R1 and R3). Because the R2 group is identical in all four compounds, we cannot assess its importance. Because of limited interactions with protein atoms, the hydroxyl group in R4 probably has limited significance, although it might influence the protonation state (and/or pH dependence) of Y-3. The removal of the charged R3 sulfonate in Y-4 leaves only the hydrophobic β-ring to interact with critical polar IN residues, presumably preventing effective binding. Lack of binding by Y-1 or Y-2 similarly could be because of an R1 hydrogen-for-sulfonate substitution or the introduction of an R5 NH2 group in Y-2. Among these three choices (H, NH2, or SO3), only sulfonate clearly carries the correct charge and lies in the correct orientation to interact with IN. In contrast, the substitution of SO3 by NH2 at R3 may well be tolerated and is presently under study. Binding may be pH-dependent, so the other three compounds might be stabilized by a pH under five or over eight.
Comparison with HIV-1 IN.
Because these compounds initially were studied for their inhibitory potency against HIV-1 IN, we checked whether the binding requirements of Y-3 also are compatible with HIV-1 IN. A structural alignment of the conserved areas of both enzymes [based on HIV-1 IN Protein Data Bank code 2ITG (13)] indicates that comparable binding of Y-3 to HIV-1 IN is possible. Two ASV IN residues of particular importance for Y-3 binding, Gln-62 and Lys-119, are equivalent to HIV-1 IN Gln-62 and His-114, respectively. HIV-1 IN and ASV IN both have a flexible loop with highly conserved residues, which is involved directly in substrate contacts (31). We now show in ASV IN that Y-3 repositions some loop residues and directly contacts others. Y-3 inhibition of HIV-1 IN is to likely proceed by an equivalent mechanism.
CONCLUSIONS
This structure of an inhibitor of both HIV-1 IN and ASV IN, solved in a complex with the latter enzyme, is the first reported example of its kind. IN inhibitor assays are done in vitro using artificial substrates and may not reflect precisely in vivo potency (32, 33). It is therefore desirable to have independent confirmation of inhibitor binding specificity. In the case of Y-3, observed in vitro potency is supported by structural data; the inhibitor binds specifically in close vicinity to the active site and to the predicted nucleic acid binding site (29–31, 34). Conformations of active site residues are altered upon inhibitor binding. Specific changes, notably the rotation of the Asp-64 side chain, are pH specific and complex. Experiments are underway to clarify their potential role in inhibition. Although the influence of Y-3 binding on the conformation of the flexible loop is clear and may account for its inhibitory activity, full interpretation of its significance awaits more data on the mode of IN-substrate binding. Although the nature of inhibition cannot be fully established, some possibilities can be ruled out. For example, there is no interference with the binding of an essential divalent cation.
Comparison of the isolated catalytic core domains of HIV-1 IN (13, 35) and ASV IN (14) indicates that, as crystallized, only the conformation of the catalytic residues of the latter is compatible with metal binding. This result was puzzling given the overall homology of these enzymes, and the known ability of HIV-1 IN to bind cations in solution (20). We now find that under some conditions of low pH, ASV IN adopts an inactive conformation incapable of binding cations (unpublished data). A similar shift could explain the apparent discrepancy in HIV-1 IN if conformational flexibility of the active site resulted in interconversion between active and inactive forms of the enzyme. Because these results were obtained from a single crystal form of the ASV IN core domain, the question remains whether IN in solution, particularly full-length IN, interacts identically with Y-3. Given that binding is only seen for the strongest of four potential inhibitors, we believe these results also are likely to hold for core domain and full-length INs in solution.
Our goal in working with this class of inhibitors was to provide leads to potential drugs targeting HIV-1 IN and a better understanding of the location of crucial target residues in IN. We have shown that Y-3, an HIV-1 IN inhibitor, stably binds to crystallized ASV IN. By demonstrating the mode of Y-3 binding to ASV IN, we have ruled out cation binding site competition as a potential mechanism for this class of inhibitors. We have pinpointed several residues in the highly conserved flexible loop and active site as potential targets for IN inactivation. This demonstrates that ASV IN can provide insight into the interaction between HIV-1 IN and Y-3.
Acknowledgments
We thank Sue Fox, Anne Arthur, and Joy Sabl for their editorial comments. Research was sponsored in part by the National Cancer Institute, Department of Health and Human Services, under contract with ABL. The contents of this publication do not necessarily reflect the views or policies of the Department of Health and Human Services nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
ABBREVIATIONS
- ASV
avian sarcoma virus
- IN
integrase
- HIV-1
HIV type 1
- ED
electron density
Footnotes
Data deposition: The coordinates of ASV IN complexed to Y-3 at pH 5.6 (code 1a5w), pH 7.5 (code 1a5x), and pH 7.5 with Mn2+ bound (code 1a5v), and the corresponding x-ray structure factors, have been deposited with the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973.
Enzyme Classification: Integrase, EC 2.7.7.-, peptide P03354 (Swiss-Prot Protein Sequence Data Bank, Dept. Biochem Med., Centre Medical Universitaire, Geneva, and European Molecular Biology Laboratory, Heidelberg) of the Gag-Pol polyprotein, refer to reverse transcriptase (RNA-directed DNA polymerase, EC 2.7.7.49).
References
- 1.Katz R A, Skalka A M. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 2.Vink C, Groeneger O A M, Plasterk R H. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goff S P. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 4.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. Structure. 1996;4:89–96. doi: 10.1016/s0969-2126(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 5.Bujacz G, Alexandratos J, Wlodawer A, Merkel G, Andrake M, Katz R A, Skalka A M. J Biol Chem. 1997;272:18161–18168. doi: 10.1074/jbc.272.29.18161. [DOI] [PubMed] [Google Scholar]
- 6.Lee S P, Han M K. Biochemistry. 1996;35:3837–3844. doi: 10.1021/bi952056p. [DOI] [PubMed] [Google Scholar]
- 7.Zheng R, Jenkins T M, Craigie R. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S P, Xiao J, Knutson J R, Lewis M S, Han M K. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- 9.Khan E, Mack J P, Katz R A, Kulkosky J, Skalka A M. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lodi P J, Ernst J, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 11.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 12.Eijkelenboom A P, Lutzke R A, Boelens R, Plasterk R H, Kaptein R, Hård K. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 13.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 14.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. J Mol Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 15.Thomas M, Brady L. Trends Biotechnol. 1997;15:167–172. doi: 10.1016/S0167-7799(97)01016-0. [DOI] [PubMed] [Google Scholar]
- 16.Neamati N, Sunder S, Pommier Y. Drug Discovery Today. 1997;2:487–498. [Google Scholar]
- 17.Nicklaus M C, Neamati N, Hong H, Mazumder A, Sunder S, Chen J, Milne G W, Pommier Y. J Med Chem. 1997;40:920–929. doi: 10.1021/jm960596u. [DOI] [PubMed] [Google Scholar]
- 18.Milne G W, Nicklaus M C, Driscoll J S, Wang S, Zaharevitz D W. J Chem Inf Comput Sci. 1994;34:1219–1224. doi: 10.1021/ci00021a032. [DOI] [PubMed] [Google Scholar]
- 19.Muzic F. Collect Czech Chem Commun. 1965;30:559–572. [Google Scholar]
- 20.Pommier Y, Pilon A A, Bajaj K, Mazumder A, Neamati N. Antivir Chem Chemother. 1997;8:463–483. [Google Scholar]
- 21.Chow S A, Vincent K A, Ellison V, Brown P O. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 22.Kulkosky J, Katz R A, Merkel G, Skalka A M. Virology. 1995;206:448–456. doi: 10.1016/s0042-6822(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Brunger A T. Nature (London) 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 25.Brunger A. x-plor A System for X-ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 26.Jones T A, Kjeldgaard M. O – The Manual. Uppsala, Sweden: Uppsala Univ.; 1994. [Google Scholar]
- 27.Sack J S. J Mol Graphics. 1988;6:244–245. [Google Scholar]
- 28.Burley S K, Petsko G A. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 29.Engelman A, Hickman A B, Craigie R. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutzke R A, Vink C, Plasterk R H. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heuer T S, Brown P O. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- 32.Farnet C M, Wang B, Lipford J R, Bushman F D. Proc Natl Acad Sci USA. 1996;93:9742–9747. doi: 10.1073/pnas.93.18.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hazuda D, Felock P, Hastings J, Pramanik B, Wolfe A, Goodarzi G, Vora A, Brackmann K, Grandgenett D. J Virol. 1997;71:807–811. doi: 10.1128/jvi.71.1.807-811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenkins T M, Esposito D, Engelman A, Craigie R. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bujacz G, Alexandratos J, Zhou-Liu Q, Clement-Mella C, Wlodawer A. FEBS Lett. 1996;398:175–178. doi: 10.1016/s0014-5793(96)01236-7. [DOI] [PubMed] [Google Scholar]
- 36.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]