Abstract
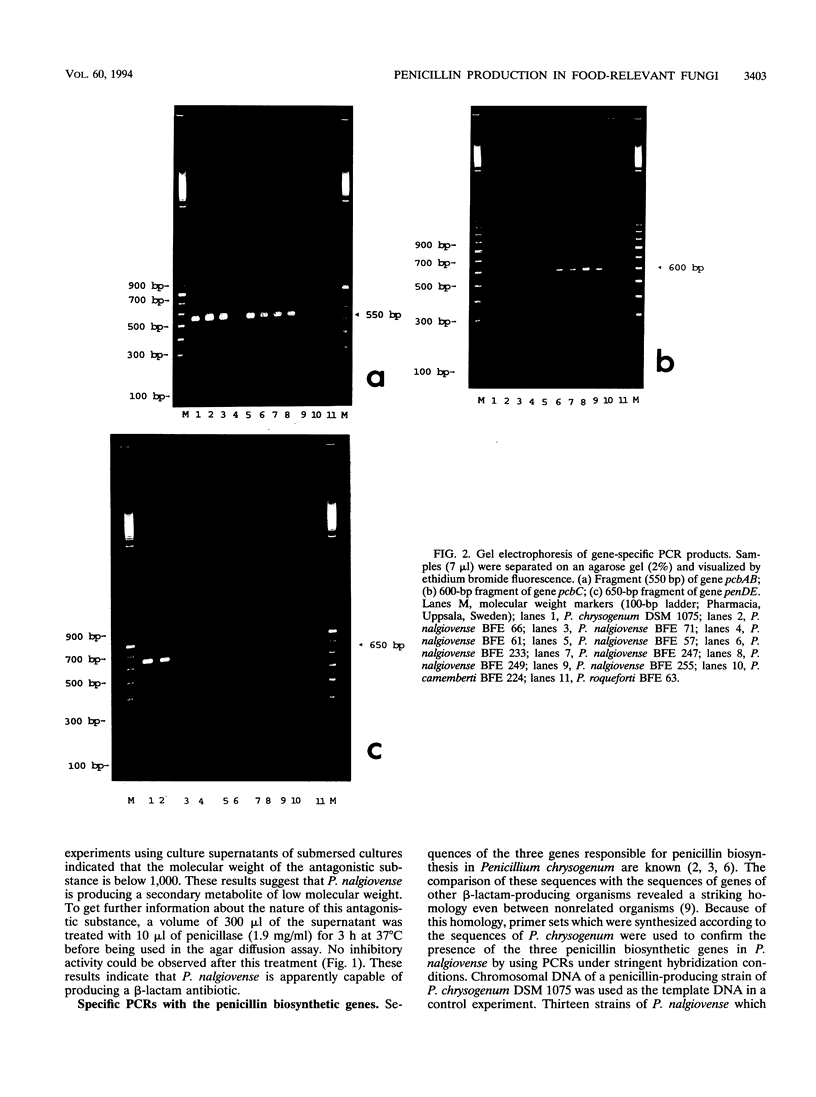
Defined strains of the genus Penicillium used as starter cultures for food and strains isolated from mold-fermented foods were analyzed for their ability to inhibit the growth of Micrococcus luteus DSM 348 used as an indicator organism. Most of the strains belonging to the species Penicillium nalgiovense showed antagonistic activity in agar diffusion assays. Penicillium camemberti and Penicillium roqueforti strains proved to be inactive in these tests. The inhibitory substance excreted by P. nalgiovense strains was totally inactivated when treated with beta-lactamase (penicillinase), indicating that a beta-lactam antibiotic is produced by these strains. This observation was verified by PCRs with primer sets specific to the [delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine] synthetase gene (pcbAB), the isopenicillin-N-synthase gene (pcbC), and the acyl coenzyme A:6-aminopenicillanic acid acyltransferase gene (penDE) from Penicillium chrysogenum using chromosomal DNA of the fungal strains as a template. These results indicate that penicillin biosynthesis is a characteristic often found in strains of P. nalgiovense. No specific PCR signal could be identified with DNA from P. camemberti and P. roqueforti.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aharonowitz Y., Cohen G., Martin J. F. Penicillin and cephalosporin biosynthetic genes: structure, organization, regulation, and evolution. Annu Rev Microbiol. 1992;46:461–495. doi: 10.1146/annurev.mi.46.100192.002333. [DOI] [PubMed] [Google Scholar]
- Barredo J. L., Cantoral J. M., Alvarez E., Díez B., Martín J. F. Cloning, sequence analysis and transcriptional study of the isopenicillin N synthase of Penicillium chrysogenum AS-P-78. Mol Gen Genet. 1989 Mar;216(1):91–98. doi: 10.1007/BF00332235. [DOI] [PubMed] [Google Scholar]
- Barredo J. L., van Solingen P., Díez B., Alvarez E., Cantoral J. M., Kattevilder A., Smaal E. B., Groenen M. A., Veenstra A. E., Martín J. F. Cloning and characterization of the acyl-coenzyme A: 6-aminopenicillanic-acid-acyltransferase gene of Penicillium chrysogenum. Gene. 1989 Nov 30;83(2):291–300. doi: 10.1016/0378-1119(89)90115-7. [DOI] [PubMed] [Google Scholar]
- Díez B., Gutiérrez S., Barredo J. L., van Solingen P., van der Voort L. H., Martín J. F. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem. 1990 Sep 25;265(27):16358–16365. [PubMed] [Google Scholar]
- Walz M., Kück U. Targeted integration into the Acremonium chrysogenum genome: disruption of the pcbC gene. Curr Genet. 1993 Nov;24(5):421–427. doi: 10.1007/BF00351851. [DOI] [PubMed] [Google Scholar]
- Yelton M. M., Hamer J. E., Timberlake W. E. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]