Abstract
The AGLCL Epstein–Barr virus (EBV) growth-transformed cell line is incapable of inducing tumors in nude mice. When the cells were transfected with a 1.3-kb CATR1 antisense cDNA construct, progressively growing lymphomas could be induced in nude mice. Chromosome analysis of the parental, transfected, and tumor cells revealed that a chromosomal translocation t(8;14)(q24.1;q32) had occurred in the transfected cells and was retained in cells derived from tumors. Moreover, enhanced c-myc expression, usually associated with this translocation, was either unchanged or under-expressed. These data suggest that the malignant transformation of the EBV growth-transformed cells was independent of c-myc expression and suggest that the CATR1 gene may act synergistically with the chromosomal translocation facilitating the conversion of AGLCL cells from a growth-transformed state to a malignant phenotype.
Epstein–Barr virus (EBV) genome positive B-cell lines have been established and characterized from Burkitt’s lymphoma (BL), and from peripheral blood leukocytes obtained from EBV seropositive individuals. Non-BL-derived EBV genome positive B cell lines are designated lymphoblastoid cell lines (LCLs) (1). Other LCLs can be obtained following infection of B-lymphocytes in vitro with transforming strains of EBV (e.g., B95-8), which leads to growth transformation of the B lymphocytes (2–4).
It has been suggested that LCLs have undergone the first of one or more initiation events in the malignant process (5). That is to say, when these cells have acquired the property to grow indefinitely in vitro and can grow in soft agar, they have not progressed to the point where the cellular events required for malignant conversion have occurred, which are defined as the ability to produce progressively growing lymphoid tumors in nude mice.
Although much information has been accumulated concerning the expression and regulation of the EBV genome in LCL and BL-derived cell lines, the mechanisms whereby EBV is involved in the development of BL is still unclear. BL-derived tumor cell lines will usually induce tumors in nude mice within 4 weeks after injection and are easily cloned in soft agar (1). The EBV-transformed LCLs (which have not been cultured for long periods of time in vitro) do not readily grow in soft agar and may occasionally induce tumors in nude mice that have limited growth potential and which subsequently regress. Other differences between BL tumor cell lines and LCLs are that the BL cell lines are usually monoclonal in origin (6, 7), are generally aneuploid (1), and have chromosomal rearrangements leading to c-myc activation and over expression (8, 9), whereas the LCLs are polyclonal and have diploid or near diploid karyotypes, particularly in early passage (1).
It has been shown that BL tumors have characteristic chromosomal translocations involving chromosomes 8 and 14, 2, or 22; the latter three encode for Igs (8, 10, 11). The chromosomal translocations that are characteristic of BL tumor cells involve the transposition of the c-myc oncogene located on chromosome 8 to a site adjacent to the IgH gene on chromosome 14 or to the κ or λ genes on chromosomes 2 or 22, respectively (8, 10, 11). The consistent finding of c-myc over expression in BL cell lines has led to the general belief that c-myc activation in B cells plays an essential role in their conversion to a malignant phenotype (12). It is not clear whether the genotypic changes necessary to convert nonmalignant EBV genome positive growth-transformed LCLs to fully malignant cells are solely confined to ploidy, chromosomal changes, or other factors yet to be determined. However, it has been demonstrated that the treatment of EBV genome positive LCLs with chemical carcinogens can result in the conversion of these cells to malignant cells capable of inducing tumors in nude mice (13, 14).
In previous studies from our laboratory, we reported on the conversion of human cells, which were derived from squamous cell carcinomas (SCC) (unable to induce tumors in nude mice), to malignant cells following methyl methanesulfonate (MMS) or N-methyl-N-nitro-N-nitrosoguanidine (MNNG) treatment (15, 16). The conversion did not appear to be related to either the frequency or location of mutations in c-myc, H-ras, Ki-ras (17), or p53 genes (18). Transfection of either a cDNA expression library or a 1.3-kb antisense CATR1 sequence (19–21) into nontumorigenic cells resulted in the conversion of these transfected cells into malignant cells (20), providing evidence that the 1.3-kb CATR1 segment of the CATR1 gene was involved in these malignant conversions. Transfection of the antisense construct reduced the expression of the CATR1 gene in tumors, suggesting that the CATR1 gene acts as a tumor suppressor gene (21). The CATR1 gene is located on the long arm of chromosome 7, near the border of bands q31 and 32 (7q31-32). This region has been reported to contain tumor suppressor activity based on the finding that loss of heterozygosity at 7q31.1-32 is associated with colon, head and neck, ovary, prostate (22–26), pancreatic (27), gastric (28), and breast cancers (29). In addition, genes located on chromosome 7 have also been reported to regulate invasion and spread of T cell lymphomas (30), and a putative tumor suppressor gene located around 7q31 was shown to suppress the cytogenetic and clinical progression of myeloid leukemias and myeloblastic syndromes (31).
In the present study, we explored the role of the CATR1 gene in the induction of a malignant phenotype by transfection of either a nondirectional cDNA expression library or an antisense construct of the 1.3-kb CATR1 segment into an EBV growth-transformed, nontumorigenic LCL (AGLCL) and evaluation of tumorigenicity in nude mice.
MATERIALS AND METHODS
Cell Cultures.
The derivation of the AGLCL cell line has been reported (2). Cytogenetic analysis of the AGLCL showed that the cell line used for this study at passage 18 had a 46,XX chromosome constitution without clonal changes.
cDNA Preparation.
Antisense CATR1 expression plasmids were constructed by ligating CATR1 cDNA to the BstXI sites of the eukaryotic expression vector pRC/RSV (Invitrogen). Individual clones were verified for antisense orientation by restriction digestion patterns (19).
Transfection with Antisense 1.3-kb cDNA Construct.
Plasmid DNA was purified from the tumor expression library as described here. The supercoiled plasmids were isolated by using a CsCl gradient. Approximately 107 rapidly growing AGLCL cells were mixed with 80 μg of a SalI linearized plasmid and Lipofectin-DNA mixture. After a 16-hr incubation, washing, and a second incubation of 24 hr, the cells were transferred to a medium containing 300 μg/ml of G418. This selective medium was changed every 2–3 days for 2 weeks. The cultures produced a large population of G418-resistant cells. These populations were designated AGLCL-TR cells (transfected AGLCL cells).
Determination of Tumorigenic Potential of AGLCL-TR Cells.
The AGLCL-TR cells were injected into 3- to 4-week-old male nude mice as described (15–17). Briefly, 5 × 106 cells were injected s.c. into the flank of the mice. Three weeks after injection, tumors greater than 2.0 cm in length were excised, and the tumor cells, designated AGLCL-CATR1, were cultured.
Southern Blot Analysis.
The integration of the cDNA antisense into AGLCL-TR genomic DNA was determined by Southern blot analysis by using a vector-specific neo-resistant gene probe. Genomic DNA was isolated according to the procedures described in Sambrook et al. (32). The DNA was digested to completion with five units of EcoRI restriction enzyme per microgram of DNA. The digested DNA was electrophoresed on 0.8% agarose gels, transferred onto nylon membranes, and hybridized for Southern blot analysis (32).
Chromosomal Analysis.
Cells were cultured for 24 hr and G-banded (GTW) according to standard cytogenetic techniques (33). Twenty metaphases were analyzed from each cell culture and karyotypes were described (34).
c-myc Expression by Northern Blot Analysis and Semi-Quantitative Reverse Transcription–PCR (RT-PCR).
Northern blot analysis was carried out according to the procedure described by Lee et al. (18). The autoradiograph was first digitized by using a twain type program and then scanned subsequently by a NIH image computer program. The bands for c-myc were normalized against HPRT bands run on each lane for each gel electrophoresis. Semi-quantitative RT-PCR was carried out in the following manner. A standard curve was generated by using c-myc and hprt genes as benchmarks for generating the data used in the curves. The n values were 3 or more values for each point measured over five different dilutions. The vertical bars at each reaction point are a calculated mean value of an n of 3 plotted as the SEM calculated by using the Student t test. In this manner, we obtained a corresponding normalized ratio of c-myc to hprt expression. (Note that quantitative RT-PCR data on hprt and c-myc expression on film are available upon request from G.M.)
Changes in CATR1 Expression as Measured by RT-PCR.
This procedure employs specific primers that anneal to specific regions in the CATR1 gene; HPRT was used as a normalization standard. One microgram of RNA was dissolved in diethyl pyrocarbonate water and incubated at 42°C for 60 min in a volume of 20 μl. The reaction mixture contained 1× PCR buffer from GIBCO/BRL, 5 mM MgCl2, 100 pmol of random hexamers, 1 unit/ml of RNAsin, 1 mM each of dATP, dGTP, dCTP and dTTP, and 100 units of Superscript II reverse transcriptase. After completion of the RT reaction, the samples were preincubated at room temperature for 10 min and heated to 99°C for 5 min to terminate the reaction. Approximately 50–200 ng of cDNA from each sample was amplified by using forward and reverse primers for either HPRT or CATR1 genes in separate reactions. All pre-PCR mixtures contained 1× PCR buffer, 1.5 mM MgCl2, 0.25 μM each of forward and reverse primers for either HPRT or CATR1, 0.2 mM each of dATP, dGTP, dCTP, and dTTP, 550 ng of Taq start antibody, and 2.5 units of Taq DNA polymerase in a final volume of 50 μl.
The following primers were used: CATR1 forward primer (FP2) 5′-AGTCCACGAAGCTAACTACCTTTG-3′ and CATR1 reverse primer (NP3) 5′-GGAGTCATTTGATCTTACCCAACA-3′. Because the tm of these two primers are 60.2°C and 61.4°C, respectively, the RT reaction was optimized with regard to not only primer concentration, but also to annealing temperature. HPRT-specific primers were the same as those listed previously in the determination of c-myc expression. The size of the HPRT product was 177 bp and of the CATR1 cDNA segment, 353 bp. For detection of the CATR1 and HPRT genes, the reactants were subjected to 33 cycles of amplification at 94°C for 1 min, 57°C for 1 min, and 68°C for 1 min followed by a 10-min incubation at 72°C. These amplification conditions place the reaction in the exponential range for each target sequence. The PCR-amplified products were analyzed on a 1.8% agarose sequencing gel, and the relative amount of target gene expression was determined by ethidium bromide staining and examination under UV light.
Cloning and Sequencing of CATR1 Gene.
RT-PCR and primer-specific areas to the 1.3-kb CATR1 element were used to clone the regions of the CATR1 gene downstream and upstream of the genetic element. Multiple rounds of amplification were performed in which the nested gene-specific primer used in a subsequent round annealed to a site in the cDNA molecule upstream of the nested-gene used in a previous round of synthesis (18).
RESULTS
Induction of Tumors by Transfected AGLCL Cells.
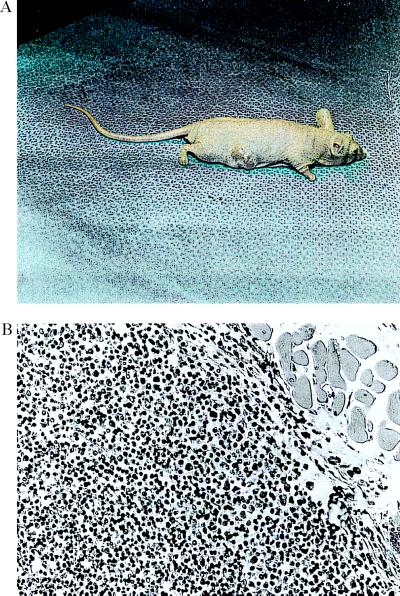
AGLCL cells (passage 18) were transfected with the 1.3-kb CATR1 cDNA antisense construct in three separate experiments followed by inoculation of the transfected cells (AGLCL-TR) into nude mice. Tumors developed within 1 week in 10 of 10 mice inoculated with the AGLCL-TR cells (Fig. 1A) and grew to ≈1.5–2.0 cm in length by 3 weeks, whereas no tumors were observed within 5 months in 7 of 7 mice inoculated with the parent AGLCL. The tumors were characterized histologically as B cell lymphomas (Fig. 1B) with sheets of large lymphoblastoid cells and minimal connective tissue. Cells were large and round, containing abundant cytoplasm and large variable shaped nuclei. The mitotic index of the tumors was 5–10%.
Figure 1.
The data in A and B present a tumor growing in the subscapular area on the back of a nude mouse. This tumor was observed 1 week after receiving a bolus of 5 × 106 cells contained in a 0.1 ml injection into this area of the back. These antisense transfected cells were recovered from four different tumors. (B) Histopathology of a tumor 3 weeks after receiving the cell bolus. The tumor was sectioned into 8-μm sections, fixed in formalin at pH 7. 0, and stained with hematoxylin/eosin. B cell lymphomas containing large lymphoblastoid cells were observed in numerous areas throughout the tumor area. The sections contained a minimal amount of connective tissue. (B, ×200.)
Detection of the 1.3-kb CATR1 cDNA Construct.
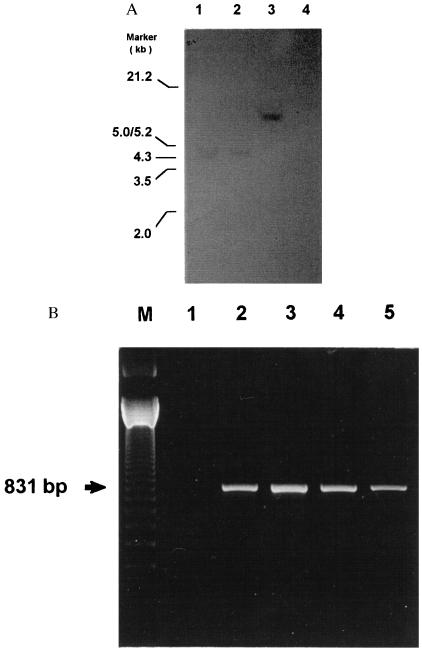
Southern blot analysis was performed on EcoRI-digested genomic DNA prepared from AGLCL-TR cells and probed with the labeled vector-specific neo-resistant gene. The transfected cells were also evaluated for transcription by RT-PCR amplification of the neo selection marker. Southern blot analysis revealed the presence of an integrated cDNA (Fig. 2A) into the genomic DNA and that the neo gene from the vector was expressed (Fig. 2B).
Figure 2.
(A) Radiograph of Southern blot analysis of genomic DNA prepared from the AGLCL 1.3-kb CATR1 cDNA antisense tranfected cells. This autoradiograph shows the integration of the vector into the genomic DNA. Each lane in the figure received 10 μg of DNA digested with EcoRI, and the membrane was probed with the labeled vector-specific neo-resistant gene probes. The arrows indicate the location on the gel of the vector DNA. Similar results were obtained by using three different transfected cell lines. The origin of the DNA is indicated at the top of each lane, i.e., lanes 1–3 were prepared from three different transfected cell lines, and lane 4 contained genomic DNA prepared from untransfected AGLCL cells. (B) Data are presented showing the RT-PCR amplification of the expressed neo selection marker. In each case 1 μg of RNA was used for the RT reaction. Oligo(dT) was used as the first strand primer in the reaction. One-tenth of the RT reaction mixture was used for PCR amplification by using primers for the Neo-1 and Neo-2 genes. These primers would amplify a 831-bp neo-specific product.
Chromosome Analysis.
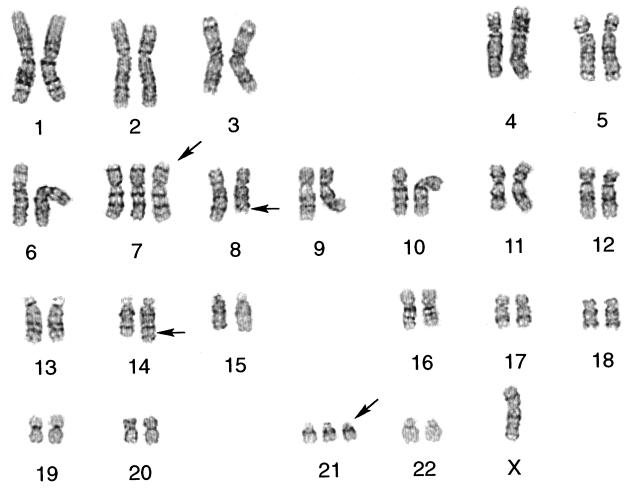
Chromosomal analysis of the control AGLCL, AGLCL-TR, and cells derived from a tumor (AGLCL-CATR1) was performed. Chromosome analysis of the AGLCL-TR cells revealed three related clones, which shared the characteristic BL t(8;14)(q24.1;q32) translocation and loss of an X chromosome (Fig. 3). Clonal evolution was demonstrated by the acquisition of a +21 chromosome by a second clone, and by +7 and +21 chromosomes by a third clone. Karyotypic instability was also a feature of these transfected cells as indicated by additional nonclonal changes in one cell line. To clarify the changes observed in the chromosome complementation of the cells in different stages of progression, we offer the following evidence. The parental AGLCL, the EBV growth transformed transfected AGLCL cells prior to injection of the tranfected cells into the surrogate nude mouse, showed no clonal chromosome abnormalities. Moreover, within the population of AGLCL-TR cells there were three cytogenetically related clones with the t(8;14) common to all. The AGLCL-CATR1 5-10-9 cells showed a single dominant clone with −X,+7 and +21 in addition to the t(8;14) which represented, cytogenetically speaking, the most clonally evolved line found in the AGLCL-TR cells. The AGLCL-CATR1 5–19-9 cells demonstrated the following clonal karyotype: 47,X,−X,+7,t(8;14)(q24.1;q;32),+21. Three other cells, related to the clone, showed other nonclonal changes identified here as 47,X,−X,+add(7)(p22),t(8;14)(q24.1;q32),+21[1]/47,X,−X,+7,t(8;14)(q24.1;q32),+21,−22,+mar[1]/48,X,−X,+7,+11,t(8;14)(q24.1;q32),+21[1]. Interestingly, the AGLCL-CATR cells represented a single clone containing the −X,+7,t(8;14)(q24.1;q32) and +21 as observed in the AGLCL-TR cells with the greatest clonal evolution. This finding suggests that this clone had a survival advantage in the nude mouse. The translocations t(8;14), t(2;8) and t(8;22) have consistently been shown to result in deregulation of the transcription of the c-myc oncogene resulting in overexpression in lymphomas and leading to speculation that the translocations are involved in malignant transformation mainly through deregulation of c-myc transcription (35).
Figure 3.
G-banded karyotype of AGLCL-CATR1 antisense transfected cell line prepared from a CA nude mouse tumor. The cell line is designated 5-1-9-9 that exhibits clonal changes including loss of an X chromosome, trisomy 7,t(8;14)(q24.1;q32), and trisomy 21: 47,X,−X,7,t(8;14)(q24.1;q32),+21.
Expression of c-myc as Analyzed by Northern Blot Analysis in Tumors.
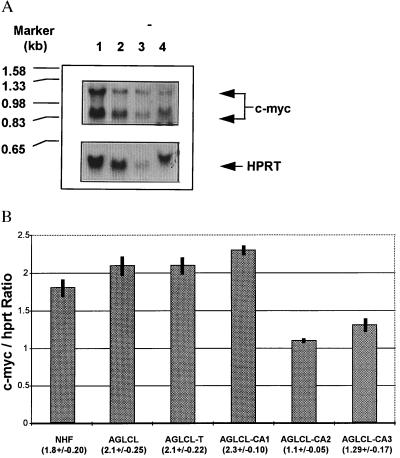
In contrast to the results of others, there was no significant increase in c-myc expression in AGLCL-CATR transfected cell-induced tumors. In fact, the results presented in Fig. 4 show that c-myc expression was not found to increase. Moreover, in tumor cell lines the expression level of c-myc was not increased measurably over the expression level in nontransfected parent AGLCL line or in three different other cell lines isolated from the transfected tumors, (Fig. 4A, see data presented in lane 2 and 3; and see data presented for quantitative RT-PCR in Fig. 4B).
Figure 4.
(A) Northern blot analysis of levels of c-myc expression were compared between the parental nontransfected AGLCL cells and AGLCL-transfected cell-induced tumors. Lanes: 1, results of expression of the AGLCL-transfected induced tumors; 2 and 3, data on expression from cell lines derived from tumors induced by different transfected AGLCL cells; 4, data on parental AGLCL cells before transfection. Each lane contained 20 μg of total RNA. The arrows (Upper) indicate where the c-myc probe detected levels of expression in each of the cell lines and tumor. (Lower) HPRT probe was used as an internal control for expression in all the preparations. (B) Digital analysis of the semi-quantitative PCR results revealed that c-myc levels of expression, when normalized to internal hprt levels, yielded data that indicated that there was no increase in levels of c-myc expression. The values in parentheses are the mean values for three measurements ± SD. NHF, cells prepared from normal human neonatal foreskins; AGLCL, parental EBV immortalized LCL cells; AGLCL, designate transfected-induced AGLCL solid tumor-derived cell lines AGLCL-CA1, AGLCL-CA2, and AGLCL-CA3. The numerical values presented under the bar graphs represent 6 samples for 5 dilutions at 23 cycles for both c-myc and hprt in the Perkin–Elmer Cycler.
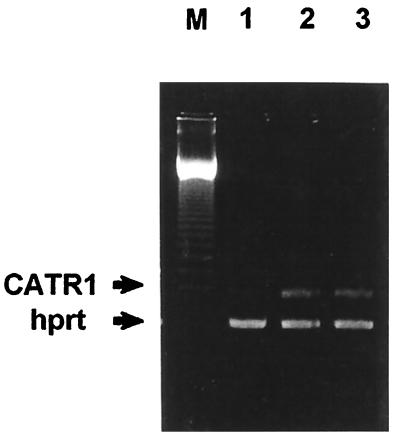
Expression of CATR1 in Tumors.
When these cells were examined by semi-quantitative RT-PCR, the tumors induced by the AGLCL-TR cells showed greater than 90% reduction in the level of CATR1 expression (Fig. 5) when compared with either the parental AGLCL cells or normal human fibroblasts (15). The RT-PCR data gathered for the c-myc gene confirmed the Northern blot analysis data, where we observed no detectable change in the level of c-myc expression by this procedure either. In addition, the data in Fig. 4A shows that CATR1 mRNA is not expressed in the AGLCL-CATR1 cells, but is clearly detectable and present in AGLCL parent cells and in normal human fibroblasts. These data suggested that CATR1 may be acting as a tumor suppressor gene [GenBank accession no. U25433 update (14)].
Figure 5.
The cell types evaluated here are presented in the legend of Fig. 1. The PCR products were separated on an 1.8% agarose gel and stained with ethidium bromide. A 123-bp DNA ladder was used as a marker.
DISCUSSION
We have previously demonstrated that cells derived from human SCC, that were nontumorigenic in nude mice, could be converted to a tumorigenic phenotype by treatment with MMS or MNNG (15, 16). From tumors induced by these treated cells, we isolated a putative tumor suppressor gene, CATR1, and demonstrated that transfection of SCC or chemically transformed human foreskin cells (with either a nondirectional cDNA expression library and/or subsequently with a 1.3-kb CATR1 antisense cDNA construct) will convert these cells to a malignant phenotype (20). These cells will induce growing tumors in nude mice within 1–3 weeks. We have also examined 12 head and neck tumors and found that the expression of the CATR1 gene is dramatically reduced in all samples (unpublished data). In control, nontumorigenic tissues, the level of expression varies but is consistently present at levels above that detected in tumors.
Translocations between chromosomes 8 and 14 t(8;14) have predominantly been found in EBV-associated BLs (6, 7) with variants having translocations between chromosomes 8 and 2 or 22 [t(2;8) or t(8;22)]. The consistent appearance of these translocations in BL, which transposes the c-myc gene from chromosome 8 to a position adjacent to the IgH gene on chromosome 14 or inserts the kappa or lambda genes from chromosomes 2 or 22 distal (3′) to the c-myc oncogene on chromosome 8 (36, 37), strongly suggests that the translocations are an essential component of BL development.
The translocations t(8;14), t(2;8), and t(8;22) have consistently been shown to result in deregulation of the transcription of the c-myc oncogene resulting in overexpression of c-myc in lymphomas and leading to the hypothesis that the translocations are involved in malignant transformation through deregulation of c-myc transcription (8, 33–38). To determine the involvement of c-myc in the tumorigenic conversion of the EBV growth-transformed AGLCLs, c-myc expression in three tumors produced by the AGLCL-TR cells were evaluated and compared with expression of c-myc in the parental AGLCL cells and in normal human fibroblasts. In contrast to the results of other studies, there was no significant increase in c-myc expression in AGLCL-TR cell-induced tumor cell lines. In fact, the results presented in Fig. 4A show that c-myc expression is not increased in the tumors and tumor cell lines. These data were obtained by Northern blot analysis with the same tumor specimens (Fig. 4A) and was confirmed by semi-quantitative RT-PCR (Fig. 4B).
In our studies, an 8;14 translocation t(8;14)(q24.1;q32) and other chromosomal rearrangements were observed in the AGLCL-CATR1 cells without an accompanying change in c-myc expression, providing evidence that these two events are not always linked and that the cDNA expression library induced a tumorigenic phenotype independent of c-myc regulation. This finding is similar to that observed when EBV growth-transformed LCLs were converted to tumorigenic cells by treatment with chemical carcinogens (13, 14). The treated cells showed chromosomal rearrangement but did not demonstrate amplification of the cellular oncogenes, N--ras, or H-ras, or c-myb (14). These latter findings are compatible with our previous reports that H-ras and c-myc were unaffected when nontransplantable SCC cell lines were converted to malignancy by MMS or MNNG (15, 16). It was suggested that the mechanism for malignant transformation of EBV LCL by chemical carcinogens “may involve the activation of unique transforming genes, which heretofore have not been described” (5). It is possible that CATR1 may be one of the genes responsible for the malignant conversion of EBV-transformed LCLs by chemical carcinogens. In the absence of c-myc overexpression, it seems that suppression of the CATR1 gene acts synergistically with the t(8;14) translocation to produce a phenotype that is tumorigenic in nude mice.
ABBREVIATIONS
- EBV
Epstein–Barr virus
- BL
Burkitt’s lymphoma
- LCL
lymphoblastoid cell line
- SCC
squamous cell carcinoma
- MMS
methyl methanesulfonate
- MNNG
N-methyl-N-nitro-N-nitrosoguanidine
- RT-PCR
reverse transcription–PCR
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U25433).
References
- 1.Nilsson K. In: The Epstein Barr Virus. Epstein M A, Achong B G, editors. New York: Springer; 1979. pp. 225–281. [Google Scholar]
- 2.Glaser R, Theil K, Bonneau R H, Sheridan J F, Vasquez M, Allen C M. Int J Cancer. 1993;55:281–287. doi: 10.1002/ijc.2910550219. [DOI] [PubMed] [Google Scholar]
- 3.Glaser R, Tarr K L, Dangel A W. Int J Cancer. 1989;44:95–100. doi: 10.1002/ijc.2910440118. [DOI] [PubMed] [Google Scholar]
- 4.Zech L, Haglund U, Nilsson K, Klein G. Int J Cancer. 1976;17:47–56. doi: 10.1002/ijc.2910170108. [DOI] [PubMed] [Google Scholar]
- 5.Henderson E E. J Natl Cancer Inst. 1988;80:476–483. doi: 10.1093/jnci/80.7.476. [DOI] [PubMed] [Google Scholar]
- 6.Mosier D E, Gulizia R J, Baird S M, Wilson D B. Nature (London) 1988;335:256–259. doi: 10.1038/335256a0. [DOI] [PubMed] [Google Scholar]
- 7.Fialkow P J, Klein E, Klein G, Clifford P, Singh S. J Exp Med. 1973;138:89–102. doi: 10.1084/jem.138.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein G, Klein E. Cancer Res. 1986;46:3211–3224. [PubMed] [Google Scholar]
- 9.Rabbitts T H, Hamlyn P H, Baer R. Nature (London) 1983;306:760–765. doi: 10.1038/306760a0. [DOI] [PubMed] [Google Scholar]
- 10.Pelicci P G, Knowles D M, Magrath I, Dalla-Favera R. Proc Natl Acad Sci USA. 1986;83:2984–2988. doi: 10.1073/pnas.83.9.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo R C, Croce C M. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caccia N C, Mak T W, Klein G. J Cell Physiol. 1984;3:199–208. doi: 10.1002/jcp.1041210423. [DOI] [PubMed] [Google Scholar]
- 13.Le Francois-Chabas D, Montagnier L. CRS Acad Sci Ser III. 1983;296:283–286. [PubMed] [Google Scholar]
- 14.Kessler D J, Heilman C A, Cossman J, Maguire R T, Thorgeirsson S S. Cancer Res. 1987;471:527–531. [PubMed] [Google Scholar]
- 15.Milo G E, Shuler C F, Stoner G, Chen J C. Cell Biol Toxicol. 1992;8:193–205. doi: 10.1007/BF00156730. [DOI] [PubMed] [Google Scholar]
- 16.Shuler C, Kurian P, French B T, Noyes I, Sital N, Hollering J, Trewyn R W, Schuller D, Milo G E. Teratogen Carcinogen Mutagen. 1990;10:53–65. doi: 10.1002/tcm.1770100107. [DOI] [PubMed] [Google Scholar]
- 17.Milo G E, Shuler C, Kurian P, French B T, Mannix D G, Noyes I, Hollering J, Sital N, Schuller D, Trewyn R W. Proc Natl Acad Sci USA. 1990;87:1268–1272. doi: 10.1073/pnas.87.4.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H, Li D, Prior T, Casto B C, Weghorst C, Shuler C, Milo G E. Cell Biol Toxicol. 1997;13:419–434. doi: 10.1023/a:1007419810705. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Noyes I, Shuler C, Milo G E. Proc Natl Acad Sci USA. 1995;92:6409–6413. doi: 10.1073/pnas.92.14.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milo G E, Li D, Casto B C, Theil K, Shuler C, Noyes I, Chen J. Proc Natl Acad Sci USA. 1996;93:5229–5234. doi: 10.1073/pnas.93.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Yan H, Chen J, Casto B C, Theil K S, Milo G E. Carcinogenesis. 1996;17:1751–1755. doi: 10.1093/carcin/17.8.1751. [DOI] [PubMed] [Google Scholar]
- 22.Zenklusen J C, Bieche I, Lidereau R, Conti J C. Proc Natl Acad Sci USA. 1994;91:12155–12158. doi: 10.1073/pnas.91.25.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zenklusen J C, Thompson J C, Klein-Szanto A J, Conti C J. Cancer Res. 1995;55:1347–1350. [PubMed] [Google Scholar]
- 24.Zenklusen J C, Weitzel J N, Ball H G, Conti C J. Oncogene. 1995;11:359–363. [PubMed] [Google Scholar]
- 25.Zenklusen J C, Thompson J C, Troncoso P, Kagan J, Conti C J. Cancer Res. 1994;54:6370–6373. [PubMed] [Google Scholar]
- 26.Oakahashi S, Shan A L, Ritland S R, Delacey K A, Bostwick D G, Lieber M M, Thibodeau S N, Jenkins R B. Cancer Res. 1995;55:4114–4119. [PubMed] [Google Scholar]
- 27.Achille A, Biasi M O, Zamboni G, Bogina G, Magalini A R, Pederzoli P, Perucho M, Scarpa A. Cancer Res. 1996;56:3808–3813. [PubMed] [Google Scholar]
- 28.Kuniyasu H, Yasui W, Yokozaki H, Akagi M, Akama Y, Kitahara K, Fujii K, Tahara E. Int J Cancer. 1994;59:597–600. doi: 10.1002/ijc.2910590504. [DOI] [PubMed] [Google Scholar]
- 29.Bieche I, Champeme M H, Matifas F, Hacene K, Callahan R, Lidereau R. Lancet. 1992;339:139–143. doi: 10.1016/0140-6736(92)90208-k. [DOI] [PubMed] [Google Scholar]
- 30.Collard J G, van de Poll M, Scheffer A, Roos E, Hopman A H, Geurts van Kessel A H, van Dongen J J. Cancer Res. 1987;47:6666–6670. [PubMed] [Google Scholar]
- 31.Pedersen B, Ellegaard J. Cancer Genet Cytogenet. 1994;78:181–188. doi: 10.1016/0165-4608(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 9.31–9.58. [Google Scholar]
- 33.Gustashaw K. In: The ACT Cytogenetics Laboratory Manual. 2nd Ed. Barch M J, editor. New York: Raven; 1991. p. 222. [Google Scholar]
- 34.Mitelman F. An International System for Human Cytogenetic Nomenclature. Basel: Karger; 1995. , Part 2, pp. 5–12. [Google Scholar]
- 35.Nishikura K, Erikson J, ar-Rushdi A, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1985;82:2900–2904. doi: 10.1073/pnas.82.9.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croce C M, Thierfelder W, Erikson J, Nishikura K, Finan J, Lenoir G M, Nowell P C. Proc Natl Acad Sci USA. 1983;80:6922–6926. doi: 10.1073/pnas.80.22.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erikson J, Finan J, Nowell P C, Croce C M. Proc Natl Acad Sci USA. 1982;79:5611–5615. doi: 10.1073/pnas.79.18.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein G, Klein E. Immunol Today. 1985;6:208–215. doi: 10.1016/0167-5699(85)90036-2. [DOI] [PubMed] [Google Scholar]