Abstract
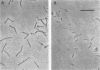
A bacterium capable of anaerobic growth via reductive dehalogenation of 2-chlorophenol was isolated from a culture enriched from sediment taken from a small stream near Lansing, Mich. The organism, designated strain 2CP-1, is a gram-negative rod ca. 3 by 0.5 micron in size and is a catalase-negative, oxidase-negative, facultative anaerobe that forms small red colonies in anaerobic media. The organism grew in reduced anaerobic mineral medium supplemented with 2-chlorophenol, acetate, and vitamins, producing phenol as a product. It did not grow when either 2-chlorophenol or acetate was omitted. The growth yield was about 3 g of protein per mol of 2-chlorophenol dechlorinated, and the doubling time was 3.7 days. Only the ortho position was dehalogenated, and additional chlorines at other positions decreased or blocked ortho dechlorination. The organism also grew with fumarate as its electron acceptor. Dechlorination activity is inducible, since cultures grown in fumarate containing medium with 2-chlorophenol rapidly dechlorinated additional 2-chlorophenol, while cultures grown in the same medium without 2-chlorophenol did not. Analysis of the organism's 16S rRNA sequence revealed that it is a member of the delta proteobacteria, more closely related to the myxobacteria than to the sulfidogenic bacteria.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990 Jun;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfing J. Reductive dechlorination of 3-chlorobenzoate is coupled to ATP production and growth in an anaerobic bacterium, strain DCB-1. Arch Microbiol. 1990;153(3):264–266. doi: 10.1007/BF00249079. [DOI] [PubMed] [Google Scholar]
- Häggblom M. M. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol Rev. 1992 Sep;9(1):29–71. doi: 10.1111/j.1574-6968.1992.tb05823.x. [DOI] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Giovannoni S. J., White D. C., Champine J. E., Phillips E. J., Gorby Y. A., Goodwin S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159(4):336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- Madsen T., Licht D. Isolation and characterization of an anaerobic chlorophenol-transforming bacterium. Appl Environ Microbiol. 1992 Sep;58(9):2874–2878. doi: 10.1128/aem.58.9.2874-2878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn W. W., Kennedy K. J. Reductive dehalogenation of chlorophenols by Desulfomonile tiedjei DCB-1. Appl Environ Microbiol. 1992 Apr;58(4):1367–1370. doi: 10.1128/aem.58.4.1367-1370.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn W. W., Tiedje J. M. Microbial reductive dehalogenation. Microbiol Rev. 1992 Sep;56(3):482–507. doi: 10.1128/mr.56.3.482-507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton D. R., Tiedje J. M. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic Acid. Appl Environ Microbiol. 1984 Oct;48(4):840–848. doi: 10.1128/aem.48.4.840-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T. O., Linkfield T. G., Tiedje J. M. Physiological characterization of strain DCB-1, a unique dehalogenating sulfidogenic bacterium. Appl Environ Microbiol. 1988 Dec;54(12):2938–2943. doi: 10.1128/aem.54.12.2938-2943.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suflita J. M., Horowitz A., Shelton D. R., Tiedje J. M. Dehalogenation: a novel pathway for the anaerobic biodegradation of haloaromatic compounds. Science. 1982 Dec 10;218(4577):1115–1117. doi: 10.1126/science.218.4577.1115. [DOI] [PubMed] [Google Scholar]
- Utkin I., Woese C., Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994 Oct;44(4):612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- WOLIN E. A., WOLIN M. J., WOLFE R. S. FORMATION OF METHANE BY BACTERIAL EXTRACTS. J Biol Chem. 1963 Aug;238:2882–2886. [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wiegel J. Sequential anaerobic degradation of 2,4-dichlorophenol in freshwater sediments. Appl Environ Microbiol. 1990 Apr;56(4):1119–1127. doi: 10.1128/aem.56.4.1119-1127.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]