Abstract
The MBNL and CELF proteins act antagonistically to control the alternative splicing of specific exons during mammalian postnatal development. This process is dysregulated in myotonic dystrophy because MBNL proteins are sequestered by (CUG)n and (CCUG)n RNAs expressed from mutant DMPK and ZNF9 genes, respectively. While these observations predict that MBNL proteins have a higher affinity for these pathogenic RNAs versus their normal splicing targets, we demonstrate that MBNL1 possesses comparably high affinities for (CUG)n and (CAG)n RNAs as well as a splicing target, Tnnt3. Mapping of a MBNL1-binding site upstream of the Tnnt3 fetal exon indicates that a preferred binding site for this protein is a GC-rich RNA hairpin containing a pyrimidine mismatch. To investigate how pathogenic RNAs sequester MBNL1 in DM1 cells, we used a combination of chemical/enzymatic structure probing and electron microscopy to determine that MBNL1 forms a ring-like structure which binds to the dsCUG helix. While the MBNL1 N-terminal region is required for RNA binding, the C-terminal region mediates homotypic interactions which may stabilize intra- and/or inter-ring interactions. Our results provide a mechanistic basis for dsCUG-induced MBNL1 sequestration and highlight a striking similarity in the binding sites for MBNL proteins on splicing precursor and pathogenic RNAs.
INTRODUCTION
To promote the assembly of functional ribonucleoprotein (RNP) complexes, RNA-binding proteins possess several different structural motifs which confer recognition of RNA sequence and structural elements (1). The RNA-recognition motif (RRM), which is the most common RNA-binding domain, consists of a βαββαβ-fold in which specific amino acids in the two central β-strands contact bases in a single-stranded (ss)RNA target site (2). Additional ssRNA-binding motifs include the KH domain, arginine-rich sequences and zinc-finger related motifs. An important example of the latter motif is found in the TTP/TIS11/ZFP36 proteins which promote the deadenylation and turnover of target mRNAs which contain a 3′ untranslated region (3′ UTR) class II AU-rich element (ARE) (3). The NMR structure of a pair of CX8CX5CX3H fingers in the TIS11d tandem zinc finger (TZF) domain protein reveals that these zinc fingers, which are separated by an 18-residue linker, bind to adjacent 5′-UAUU-3′ sequence elements (4). For double-stranded (ds)RNAs, the most common motif is the dsRNA-binding domain (dsRBD) which recognizes the A-form helical conformation (5). The dsRBD structure, which consists of a αβββα-fold with two highly conserved basic loops, interacts with successive grooves on the RNA helix (1,6). In addition, some zinc finger (ZF) proteins recognize both sequence and structural elements in their target RNAs, including TFIIIA which simultaneously contacts ssRNA and dsRNA regions in 5S RNA (7).
Previous studies have suggested that the muscleblind-like (MBNL) family of alternative splicing factors, which possess multiple copies of a TTP/TIS11-related CCCH motif (CX7CX4-6CX3H), also recognize both ssRNA and dsRNA elements (8,9). Similar to TIS11d, the CCCH motifs in MBNL proteins are organized as tandem pairs separated by a 14–16-residue linker (9–11). The MBNL proteins were first characterized as factors involved in the pathogenesis of the neuromuscular disease myotonic dystrophy (DM) (9). DM is caused by the expansion of structurally similar microsatellites in two functionally unrelated genes. Type 1 (DM1) disease is associated with (CTG)n expansions in the 3′-UTR of the DMPK gene while (CCTG)n expansions in the first intron of ZNF9 results in type 2 (DM2) disease (12). The RNA-mediated pathogenesis model for DM suggests that these expansions are toxic at the RNA level because DM1 and DM2 mutant RNAs fold into stable RNA hairpins which bind MBNL proteins with high affinity. MBNL proteins are pre-mRNA alternative splicing factors and loss of MBNL1 activity in DM tissues leads to persistence of fetal splicing patterns in the adult. In support of this model, MBNL1 binds to (CUG)n and (CCUG)n expansion RNAs (CUGexp, CCUGexp) and co-localizes with nuclear RNA (ribonuclear) foci in DM1 and DM2 skeletal muscle and brain, with a concomitant reduction in the nucleoplasmic level (13,14). Moreover, DM1 ribonuclear foci are lost following siRNA-induced downregulation of MBNL1 expression (15). Finally, Mbnl1 knockout mice recapitulate several clinical defects characteristic of DM, including myotonia and subcapsular particulate cataracts (16).
These observations indicate that a critical initiating event in the DM pathogenesis pathway is loss of MBNL function leading to defective pre-mRNA splicing. Apparently inconsistent with this view is a recent finding that several of the cardinal features of DM, including skeletal muscle myotonia and cardiac conduction defects, are recapitulated in a transgenic mouse model which overexpresses an inducible GFP-DMPK 3′-UTR transgene that contains only five CTG repeats (14). Surprisingly, these mice do not develop ribonuclear foci and the subcellular localization pattern of Mbnl1 is not altered. This study raises the question of whether foci formation is required for loss of MBNL1 function and highlights the need to further investigate the interactions of muscleblind-like proteins with both normal target and pathogenic RNAs.
Here, we report that the MBNL1 protein preferentially recognizes GC-rich RNA helices containing a pyrimidine mismatch on both normal splicing substrates and pathogenic RNAs. Furthermore, MBNL1 binds selectively to the stem region of CUGexp RNA and can be visualized as a ring-like structure in the electron microscope. This study introduces the possibility that the tandem arrangement of MBNL high affinity binding sites present on CUGexp RNA results in a stacked ring complex which effectively traps MBNL1 and inhibits its role as an alternative splicing factor during postnatal development.
MATERIALS AND METHODS
Plasmids
To construct pGEX-6P-His, a XhoI–NotI fragment encoding the His6 tag from pGEX-MBNL1 was inserted into XhoI–NotI digested pGEX-6P-1. pGEX-6P-MBNL1-His was constructed by inserting a BamHI–XhoI fragment from pGEX-MBNL1 into BamHI–XhoI digested pGEX-6P-His. The MBNL1 N-terminal region (residues 1–253) was amplified using MSS2759 and MSS2760, digested with BamHI and XhoI and inserted into pGEX-6P-His to create pGEX-6P-MBNL1-N-His. For the yeast two-hybrid system, either full length, amino terminal (residues 1–264) or carboxyl terminal (residues 239–382) MBNL1 cDNAs were PCR amplified using the following primers and inserted into pGBKT7 or pGADT7 (Clontech, Mountain View, CA, USA) at SmaI and BamHI sites: (i) full length, MSS1163 (forward primer) and MSS1166 (reverse primer); (ii) N-terminal region, MSS1164 (forward) and MSS1166 (reverse); (iii) C-terminal region, MSS1163 (forward) and MSS1165 (reverse). To create pcDNA3-V5, MSS3045 and MSS3046 were subjected to a 10-cycle PCR reaction: 94°C 30 s, 50°C 20 s, 72°C 20 s. The resulting DNA fragment was gel purified followed by digestion with NheI and BamHI, and inserted into NheI–BamHI digested pcDNA3.1(+) (Invitrogen, Carlsbad, CA, USA). The BamHI–XhoI fragment from pGEX-6P-MBNL1-His was inserted into pcDNA3-V5 at BamHI and XhoI sites to create pcDNA-V5-MBNL1. The BamHI–XhoI fragments from pGEX-6P-MBNL1-His and pGEX-6P-MBNL1-N-His were inserted into pcDNA3.1(+)/myc-His A (Invitrogen) to create pcDNA3-MBNL1-mycHis and pcDNA3-MBNL1-N-mycHis, respectively. The CUGBP1-coding sequence was PCR amplified using MSS2699 and MSS2700 and inserted into pcDNA3.1(+)/myc-His A at BamHI and XhoI sites. The Tnnt3 minigene was prepared by amplifying the mouse Tnnt3 genomic region between exons 8 and 9 using MSS1949 and MSS1950 and inserting the PCR product into pSG5(Stratagene, La Jolla, CA, USA) at the EcoRI site. The mutant Tnnt3 minigenes, pSG5-Tnnt3Δ10 and pSG-Tnnt3/gg and pSG-Tnnt3/au, were generated by site-directed mutatgenesis using MSS2129/MSS2130, MSS3131/3132 and MSS3163/MSS3164, respectively. Wild-type pSG5-Tnnt3 (100 ng) (with 125 ng of each of the primers) was subjected to the following PCR reaction: 94°C 30 s, 50°C 1 min, 72°C 8 min, 20 cycles using Pfu DNA polymerase (Stratagene). After DpnI digestion, the PCR product was transformed into DH10B and mutants were identified by plasmid DNA sequencing. Using pSG5-Tnnt3 as a template, PCR fragments generated from primer pairs MSS1865/MSS1879 and MSS1884/MSS1866 were TOPO-cloned into pCR4-TOPO (Invitrogen) to make pTOPO-T5.1 and pTOPO-T5.45, respectively.
Recombinant protein preparation
For the preparation of recombinant proteins, BL21(DE3) RP containing pGEX-6P-1-MBNL1 or pGEX-6P-1-MBNL1-N were grown to OD600 = 0.5 followed by induction with 1 mM IPTG for 2 h at 30°C. Cells were collected and resuspended in lysis buffer containing 25 mM Tris–Cl, pH 8.0, 0.5 M NaCl, 10 mM imidazole, 2 mM β-mercaptoethanol, 2 mg/ml lysozyme, 10 μg/ml DNase I, 5% glycerol, 0.1% Triton X-100 supplemented with protease inhibitors. The cell suspension was incubated on ice for 30 min with stirring prior to sonication and centrifugation at 12 000g. For protein purification, Ni-NTA-Sepharose (Amersham/GE Healthcare, Piscataway, NJ, USA) (12 ml) was incubated with the supernatant for 1 h at 4°C and washed three times with 40 ml of wash buffer containing 25 mM Tris–Cl, pH 8.0, 0.5 M NaCl, 20 mM imidazole, 0.1% Triton X-100, followed by three 10 ml elutions in 25 mM Tris–Cl, pH 8.0, 0.5 M NaCl, 250 mM imidazole, 0.1% Triton X-100. Subsequently, β-mercaptoethanol was added (10 mM final concentration) to the eluate, which was incubated with 2 ml glutathione-Sepharose (Amersham) for 1 h at 4°C. After three washes (10 ml each) of buffer (WB) containing 25 mM Tris–Cl, pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol, 0.1% Triton X-100, the glutathione-Sepharose beads were incubated with 4 ml WB containing 40 U of PreScission protease (Amersham) at 4°C overnight. The supernatant was collected following brief centrifugation and concentrated to 1–8 mg/ml.
Chemical and enzymatic analysis of RNA structures
Transcription reactions were carried out in a 50 µl volume which contained 2 μg of each DNA template, 1 mM rNTPs, 3.3 mM guanosine, 60 U of ribonuclease inhibitor RNase Out (Invitrogen), 200 U of T7 RNA polymerase (Ambion, Austin, TX, USA), 10 mM DTT, 40 mM Tris-HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl. The reaction was performed at 37°C for 2 h, the transcript was then purified on a denaturing 10% polyacrylamide gel and subsequently 5′-end-labeled with T4 polynucleotide kinase and [γ32P]ATP (3000 Ci/mmol). The labeled RNA was re-purified by electrophoresis on a denaturing 10% polyacrylamide gel. Prior to structure probing, the labeled RNA was subjected to a denaturation/renaturation procedure in a reaction buffer containing 50 mM Tris–HCl (pH 8.0), 60 mM KCl, 15 mM NaCl, 2 mM MgCl2 by heating the sample at 90°C for 1 min and slowly cooling to 25°C. The RNA sample was then mixed with either a 25-fold molar excess of recombinant MBNL1 in 50 mM Tris–HCl, pH 8.0, 60 mM KCl, 15 mM NaCl, 2 mM MgCl2, 2% glycerol, 0.5 mM DTT, 50 μg/ml BSA, or with buffer only (control) and incubated 20 min at 25°C. The final concentration of (CUG)54 was 20 nM and MBNL1 was 500 nM. Under these reaction conditions >95% of RNA was bound with protein as revealed by filter-binding assays. Additional control samples were prepared by mixing RNA with MBNL1 protein previously denatured by heating at 75°C for 2 min. Limited RNA digestion was initiated by mixing 5 µl of the RNA or RNA/protein sample (25 000 c.p.m.) with 5 µl of a probe solution containing either lead ions or ribonuclease T1 in reaction buffer. The reactions were performed at 25°C for 20 min and stopped by adding 20 volumes of 1× TE buffer followed by phenol/chloroform extraction. Precipitated RNAs were dissolved in a denaturation solution (7.5 M urea and 20 mM EDTA with dyes). To determine the cleavage sites, the products of RNA fragmentation were separated on 10% polyacrylamide gels containing 7.5 M urea, 90 mM Tris-borate buffer and 2 mM EDTA, along with the products of alkaline hydrolysis and limited T1 nuclease digestion of the same RNA. The alkaline hydrolysis ladder was generated by the incubation of the labeled RNA in formamide containing 0.5 mM MgCl2 at 100°C for 10 min. The partial T1 ribonuclease digestion of RNAs was performed under semi-denaturing conditions (10 mM sodium citrate, pH 5.0; 3.5 M urea) with 0.2 U/µl of the enzyme during incubation at 55°C for 10 min. Electrophoresis was performed at 1800 V (gel dimensions, 30/50 cm). The products of the structure probing reactions were visualized by PhosphorImaging (Storm; Molecular Dynamics, Sunnyvale, CA, USA) and analyzed by ImageQuant 5.2 (Molecular Dynamics).
Protein–protein interactions
For yeast two-hybrid analysis, pGBKT7-MBNL1 together with one of the pGADT7 constructs (pGADT7-T Antigen, pGADT7-MBNL1, pGADT7-MBNL11-N, pGADT7-MBNL1-C) were transformed into yeast strain AH109. In addition, pGBKT7-p53 and pGADT7-T Antigen (Clontech) were co-transformed into AH109 as a negative control. Double transformants were selected on SD/-Trp/-Leu plates. Expression of the myc-tagged GAL4 DNA-binding domain (BD) and HA-tagged GAL4 activation domain (AD) fusion proteins were confirmed by immunoblot analysis using mAb 9E10 and 16B12, respectively. To test for protein interactions, transformants were streaked onto SD/-Trp/-Leu/-His incubated at 30°C for 3–4 days and scored for growth.
To test for MBNL1–MBNL1 interactions in mammalian cells, HEK293T cells were transfected with 5 μg of pcDNA3-V5-MBNL1 alone as a control or 5 μg of pcDNA3-V5-MBNL1 together with either 5 μg of pcDNA3-MBNL1mycHis or pcDNA3-MBNL1-N-mycHis. Cells were harvested 20–24 h post-transfection by trypsinization followed by neutralization in media contain 10% fetal bovine serum and two washes in 50 mM Tris–HCl, pH 7.4, 150 mM NaCl. Cell pellets were resuspended in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% IGEPAL with protease inhibitors, and sonicated on ice (3 × 5 s). Cell debris was removed by centrifugation at 16 100g for 10 min at 4°C. Cleared lysates were treated with 200 μg/ml RNase A for 20 min on ice (17) followed by another 10 min centrifugation. Cleared lysates were mixed with Dynabeads coupled to Protein A (Invitrogen) precoated with rabbit anti-V5 polyclonal antibody (Novus, Littleton, CO, USA) and incubated at 4°C for 2 h. Dynabeads were washed three times with IPP150 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% IGEPAL) and once with 50 mM Tris–HCl, pH 7.4, 150 mM NaCl. Proteins were dissociated from the beads by heating at 95°C for 2 min in 1× SDS–PAGE sample buffer. Proteins (50% of the immunoprecipitated proteins, 2.5% of the input) were separated on 12.5% SDS–PAGE gels. Immunoblotting was performed using mAb 9E10 (1:1000) or mAb anti-V5 (1:1000, AbD Serotec).
Electron microscopy
The CUG136 RNA, transcribed from pBC-CTG136 plasmid (R. Osborne, University of Rochester), was incubated with MBNL1 protein at molar ratio of 1:2.5 or 1:10 in buffer containing 16 mM HEPES, 2 mM magnesium acetate, 0.16 mM EDTA, 0.4 mM DTT, 1 mM ATP, 50 mM potassium acetate and 16% glycerol for 30 min at 30°C. The resulting complexes were fixed in 0.6% glutaraldehyde for 5 min at room temperature and subsequently passed over a 2 ml column containing Bio-Gel A-5M (Bio-Rad, Hercules, CA, USA) equilibrated with 0.01 M Tris (pH 7.6) and 0.1 mM EDTA to remove free protein and fixatives. Protein–RNA enriched fractions were incubated with 2.5 mM spermidine and adsorbed on to glow charged carbon coated copper grids and dehydrated in a series of ethanol washes of 25, 50, 75 and 100% ethanol each for 5 min at room temperature. Samples were air dried prior to rotary shadow casting with tungsten. Protein–RNA complexes were visualized using a FEI Tecnai 12 electron microscope (FEI, Hillsboro, OR) at an accelerating voltage of 40 kV and images were captured using a 4K × 4K Gatan CCD camera using plate film or Gatan digital image capturing software (Gatan, Pleasanton, CA, USA). Plate film negatives were scanned using an Imacon scanner and supporting software (Imacon, Redmond, WA). Images were photographed at a magnification of 52K.
Photocrosslinking
To prepare whole cell lysates for cross-linking, HEK293T cells were grown in 10 cm plates and transfected with 10 μg of pcDNA3-CUGBP1mycHis, pcDNA3-MBNL1mycHis or pcDNA3-MBNL1-NmycHis. Cells were trypsinized and then neutralized in media containing 10% FBS followed by two additional PBS washes. Cells were then resuspended in 250 μl of 20 mM HEPES–KOH (pH 8.0), 100 mM KCl, 0.1% IGEPAL and protease inhibitors, sonicated and lysates centrifuged (13 200 r.p.m., 10 min, 4°C). Glycerol was added to the supernatants to a final concentration of 20%.
RNAs for photocrosslinking were uniformly labeled with 40 μCi each of (α-32P)-GTP and (α-32P)-UTP (800 Ci/mmol) in the presence of 0.5 mM ATP and CTP, 0.02 mM GTP and UTP. Cross-linking was performed by incubating 0.1 pmol RNA with 15 μl of HEK293T whole cell lysate in 25 μl reactions containing 16 mM HEPES–KOH (pH 8.0), 65 mM potassium glutamate, 2 mM Mg(OAC)2, 0.4 mM DTT, 0.16 mM EDTA, 20 mM creatine phosphate, 2 mM ATP and 16% glycerol (final concentration). Reactions were incubated at 30°C for 15 min, transferred to pre-chilled PCR caps on ice and photocrosslinked in Stratalinker (Stratagene, La Jolla, CA, USA) for 2.5 min (three times) with a 3 min interval between each irradiation. Samples were digested with 5 μg of RNase A for 20 min at 37°C and immunopurified using the anti-myc monoclonal antibody 9E10 pre-coated protein A Sepharose (Amersham). Purified proteins were fractionated on 12.5% SDS–PAGE gels followed by autoradiography.
Filter-binding assays
Uniformly labeled RNA was prepared as described previously (9). Calibration of the non-specific retention rate of the nitrocellulose filter was performed by incubating 0.01–0.1 nM RNA at 30°C for 30 min in BB [50 mM Tris-HCl (pH 8.0), 40 mM KCl, 20 mM potassium glutamate, 15 mM NaCl, 0.5 mM DTT, 0.05 U/ul RNasin (Promega)] followed by filtration through a Bio-Dot (BioRad) apparatus containing a sandwich of nitrocellulose (BioRad) and Hybond-N plus (Amersham) membranes followed by a single wash step with the same buffer. The membranes were UV-cross-linked, air dried and exposed to a phosphorimager screen. Non-specific retention on the nitrocellulose membrane was undetectable. Binding reactions were set up in the same buffer with 5 pM RNA and 3.13 × 10−12 M to 1.02 × 10−7 M of MBNL1/41-His, and incubated at 30°C for 30 min. Each reaction was applied to the Bio-Dot apparatus followed by one wash with binding buffer. Membranes were processed as described above and signals quantified using ImageQuant TL (Amersham). Standard deviations were calculated based on three independent experiments and apparent dissociation constants were calculated using a one-site binding model and GraphPad Prism (v3.00) software.
Gel shift assays
Gel shift assays were performed using modifications of previously described protocols (18,19). RNA was uniformly labeled with 40 μCi (α-32P)-GTP or UTP (800 Ci/mmol) in the presence of 0.5 mM ATP, 0.5 mM CTP, 0.02 mM GTP, 0.02 mM UTP and purified using a 5% denaturing gel containing 8 M urea. Prior to use, purified RNA was heated at 65°C for 5 min in 50 mM Tris–HCl (pH 8.0), 40 mM KCl, 20 mM potassium glutamate, 15 mM NaCl, 0.5 mM DTT, 0.5 U/μl SUPER-asin (Ambion) following by renaturation at RT. Reactions (20 μl) were assembled with 0.1 nM RNA and 0–256 nM protein in 50 mM Tris–HCl, pH 8.0, 40 mM KCl, 20 mM KGlutamate, 15 mM NaCl, 15% glycerol, 0.5 mM DTT, 20 μg/ml acetylated BSA. Alternatively, RNAs were heated using a higher salt concentration in the presence of magnesium (50 mM Tris–HCl (pH 8.0), 60 mM KCl, 120 mM potassium glutamate, 20 mM NaCl, 2 mM magnesium acetate, 0.5 mM DTT, 0.5 U/μl SUPER-asin (Ambion) and then reactions were performed in 50 mM Tris-HCl (pH 8.0), 60 mM KCl, 120 mM potassium glutamate, 20 mM NaCl, 2 mM magnesium acetate, 0.5 mM DTT, 20 μg/ml acetylated BSA. After incubation at 30°C for 30 min, reactions were immediately loaded onto a 4% polyacrylamide gel (80:1) containing 0.5 mM DTT and 5% glycerol which had been pre-run at 150 V for 1–2 h at 4°C. Gels were run in 0.5× TBE (pH 8.3) at 200 V for 2 h, fixed and dried prior to autoradiography.
RESULTS
MBNL1 interacts with an intronic stem-loop structure upstream of the Tnnt3 fetal exon
A recent mapping study identified several MBNL1-binding sites containing a core element within human cardiac troponin T (cTNT/TNNT2) intron 4 pre-mRNA immediately upstream of developmentally regulated exon 5 (8). Although the major MBNL1-binding sites in chicken cTNT are positioned downstream of exon 5, alignment of the human- and chicken-binding sites revealed a hexanucleotide consensus motif (5′-YGCUU/GY-3′). Neither the human- or chicken-binding sites were located in regions predicted to form secondary structures. Together with prior observations that MBNL proteins are sequestered by CUGexp and CCUGexp hairpins in ribonuclear foci in DM cells, this finding suggests that the MBNL proteins bind to both ssRNA and dsRNA structural motifs.
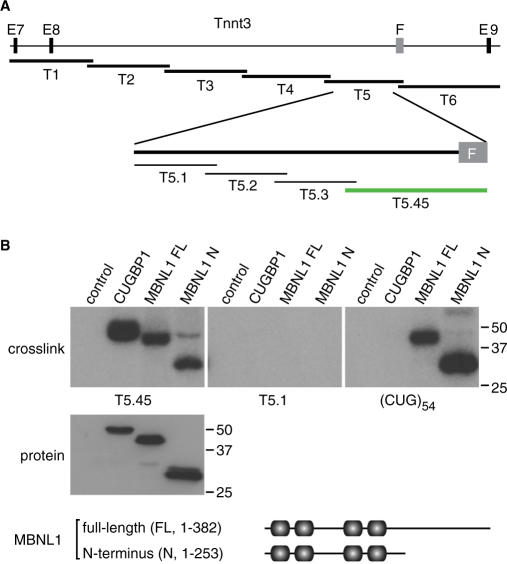
To determine if MBNL1 recognizes primarily ssRNA targets in other splicing precursors, we first mapped the MBNL1-binding site on fast skeletal muscle troponin T (Tnnt3) RNAs. This pre-mRNA was selected because earlier studies demonstrated that Tnnt3 fetal (F) exon splicing is particularly sensitive to MBNL1 levels in vitro and in vivo (16,20). Mapping was performed using a photocrosslinking protocol in which 293T cells were transfected with protein expression plasmids encoding myc-tagged versions of either CUGBP1, full-length MBNL1 (MBNL1 FL) or the MBNL1 N-terminal region (MBNL1 N) which contains the four CCCH motifs responsible for RNA binding (21). Following transfection, cell lysates were incubated with radiolabeled Tnnt3 RNAs encompassing exons 7–9 or different 500–645 nucleotide (nt) subregions designated T1-6 (Figure 1A), photocrosslinked with UV-light and digested with RNase A. Protein–RNA complexes were then immunopurified with the anti-myc monoclonal antibody (mAb) 9E10 and resolved by SDS–PAGE. Because these studies indicated that only Tnnt3 T5 RNA (500 nt) cross-linked to both MBNL1 FL and MBNL1 N proteins, this region was further subdivided into T5.1–T5.45. Interestingly, only T5.45 RNA (200 nt) cross-linked to CUGBP1 and MBNL1 proteins while T5.1 (125 nt), and the other subregions (T5.2 and T5.3, data not shown), did not (Figure 1B). In agreement with prior studies, CUGBP1 failed to cross-link to a CUGexp, (CUG)54, while both MBNL1 FL and MBNL1 N did (9).
Figure 1.
MBNL1 cross-links to both splicing precursor and pathogenic RNAs. (A) Mapping of MBNL1-binding sites on Tnnt3. Illustration shows the Tnnt3 genomic region between exons 7 (E7) and 9 (E9) (thin lines, introns; black boxes, exons) and the subregions corresponding to the RNAs tested for MBNL1 cross-linking (thick lines, T1–6; thin lines, T5.1–T5.3; green line, T5.45). (B) Interaction sites for MBNL1 and CUGBP1 on Tnnt3 pre-mRNA are positioned in a region which includes the F exon and upstream intron 8. HEK293T cells were transfected with myc-His tagged CUGBP1, MBNL1 FL or MBNL1 N followed by incubation of the corresponding cell lysates with 32P-labeled Tnnt3 T5.45, T5.1 or (CUG)54 RNAs and subsequent UV-light induced cross-linking and RNase A treatment. Labeled proteins were detected by SDS–PAGE and autoradiography (upper panels, cross-link) while protein levels in the lysates are shown by immunoblotting with the anti-myc mAb 9E10 (lower panel, protein). The primary structures of MBNL1 FL and MBNL1 N are illustrated below the immunoblot with the four CCCH motifs highlighted (shaded boxes).
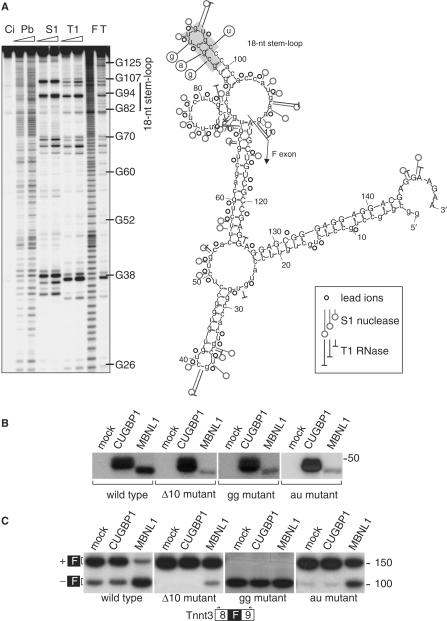
To further delineate MBNL1-binding sites on Tnnt3, we analyzed MBNL1 cross-linking to a several subregions of T5.45. One region was particularly interesting because it contains a 5′-CGCU-3′ motif which is conserved in the MBNL1-binding site in human cTNT/TNNT2 (8). Interestingly, this motif is located within a predicted 18-nt stem-loop structure (Figure 2A). We first confirmed the existence of this Tnnt3 18-nt hairpin structure using chemical and enzymatic structure probing (Figure 2A). Cross-linking assays were then used to test whether wild-type and mutant Tnnt3/98 RNA (a 98-nt subregion encompassing 83-nt of the 3′ end of intron 8 and 15-nt of the F exon) was recognized by both CUGBP1 and MBNL1 (Figure 2B). Concurrently, wild-type or mutant Tnnt3 minigenes were transfected into C2C12 cells to assay whether Tnnt3 splicing remained responsive to MBNL1 overexpression (Figure 2C). These assays were performed using C2C12 myoblasts since they showed a higher default level of Tnnt3 F exon skipping compared to 293T cells (Figure 2C, left panel, lane 1 and data not shown). Cross-linking assays confirmed that wild-type Tnnt3/98 (Figure 2B, left panel) RNA was recognized by both MBNL1 and CUGBP1. Correspondingly, the splicing pattern of the wild-type Tnnt3 minigene was not altered upon CUGBP1 overexpression (Figure 2C, left panel, compare lanes 1 and 2) while F exon exclusion was enhanced by MBNL1-mycHis (Fig. 2C, left panel, lane 3), consistent with our previous study (20).
Figure 2.
MBNL1 recognizes a RNA hairpin upstream of the Tnnt3 fetal exon. (A) Cleavage pattern (left) of the 5′-end labeled Tnnt3 151-nt transcript (a 5′ truncated form of the 200 nt T5.45 RNA) encompassing the fetal (F) exon 3′ splice site (110-nt of intron 8, 41-nt of F exon) obtained with use of three structure probes. Lanes are: Ci, incubation control or no probe added; Pb, lead ions (0.25, 0.5 mM); S1, S1 nuclease (1, 2 U/µl and 1 mM ZnCl2 was present in each reaction); T1, RNase T1 (0.5, 1 U/µl); F, formamide (statistical ladder); T, guanine-specific ladder. The sequences forming the 18-nt stem-loop structure are also indicated. Also illustrated (right) is the proposed secondary structure model of the 151-nt transcript. The cleavage sites are indicated for each probe used and the figure inset shows the probe designations and cleavage intensity classification. The F exon sequence is marked in upper case and intron 8 in lower case. The positions of the G, A and U substitutions in the 18-nt stem-loop are also indicated. (B) Photocrosslinking analysis indicates reduced MBNL1, but not CUGBP1, binding to the Tnnt3 Δ10, gg and au mutants in contrast to wild-type RNA. Photocrosslinking analysis was performed as described in Figure 1 using the same lysates (protein loading controls shown in Figure 1B) except only MBNL1 FL (MBNL1) protein was used. (C) Tnnt3 F exon skipping is impaired in the Δ10 and au mutants compared to wild type while F exon inclusion is eliminated in the gg double mutant. C2C12 cells were co-transfected with either a wild type, Δ10, gg or au point mutant splicing reporter plasmid and a protein expression plasmid for either CUGBP1mycHis or MBNL1mycHis (full-length protein only). 32P-labeled splicing products, which included (+F) or excluded (−F) the Tnnt3 F exon (black box), were detected by RT-PCR, using primers positioned in Tnnt3 exons 8 and 9 (open boxes with arrows), followed by gel electrophoresis.
Mutations in the 18-nt hairpin positioned just upstream of F exon (Figure 2A) were generated to test whether it contains an MBNL1-binding site. The relative position of this hairpin in the F exon 3′ splice site region is significant since a potential binding site for the essential splicing factor U2AF, which binds preferentially to U-rich tracts, lies just upstream of this stem-loop structure and this GC-rich hairpin contains a pyrimidine mismatch reminiscent of the structure of CUGexp and CCUGexp RNA hairpins. We first generated a 10-nt deletion within the 18-nt region to eliminate this hairpin (Figure 2A, deleted nucleotides in gray). As predicted, this mutant showed an impairment of both MBNL1 cross-linking (Figure 2B, Δ10 mutant) and F exon skipping promoted by MBNL1 overexpression (Figure 2C, Δ10 mutant). Indeed, the Δ10 deletion eliminated F exon skipping in cells transfected with the Tnnt3 minigene alone or with CUGBP1-mycHis together with the minigene (Figure 2C, Δ10 mutant, lanes 1–2) suggesting loss of MBNL1 binding and enhanced spliceosome recruitment to the F exon region.
The cross-linking and splicing results obtained with the Δ10 mutant indicated that the 18-nt hairpin was a binding site for MBNL1 and that MBNL1–hairpin interactions might promote F exon skipping. Because both sequence and structural elements could contribute to efficient MBNL1 binding, a double C→G and U→G mutant was generated that eliminated the C-C mismatch in the 18-nt hairpin, which also increases the stability of this stem-loop, and furthermore substituted a G for a U in the loop. These mutations are not predicted to alter the overall folding pattern of Tnnt3/98. This double point (gg) mutant showed considerably reduced MBNL1 cross-linking compared to wild-type Tnnt3 (Figure 2B) confirming that this region was an MBNL1-binding site. Interestingly, a similar decrease in MBNL1 cross-linking activity was also observed for the single C→G stem substitution mutant (data not shown). While loss of MBNL1 binding should promote F exon splicing, inclusion activity was completely eliminated (Figure 2C, gg mutant). Loss of F exon splicing activity could result from enhanced hairpin stability, or an increase in the purine content of the polypyrimidine (Py) region upstream of the F exon 3′ splice site, and subsequent impairment of spliceosome assembly. Interestingly, another double mutant (G→A and C→U), which preserved the predicted 18-nt stem-loop structure while reducing the GC content in the stem, also showed reduced MBNL1 cross-linking compared to wild type (Figure 2B, au mutant) while MBNL1-induced F exon skipping activity was reduced similar to the Δ10 mutant (Figure 2C, au mutant). Overall, these studies indicated that MBNL1 prefers to bind to a GC-rich stem-loop containing a pyrimidine mismatch in a normal splicing target.
Similar affinities of MBNL1 for splicing precursor and pathogenic RNAs
According to the RNA sequestration model, pathogenic RNAs outcompete normal RNA-binding targets for MBNL1 leading to loss of MBNL-mediated regulation of alternative splicing during postnatal development. However, it is not clear why MBNL1 accumulates on DM1 and DM2 expansion RNAs in ribonuclear foci. The most straightforward explanation is that MBNL1 has a higher affinity for DM pathogenic RNAs compared to its physiological RNA splicing targets. Since the cross-linking of MBNL1 to the Tnnt3 F exon 3′ splice site region and CUGexp RNAs appeared to be comparable (Figure 1B), we determined the relative affinities of MBNL1 for (CUG)54, (CAG)54 and Tnnt3/5.45 using recombinant MBNL1 protein in filter binding and gel shift assays.
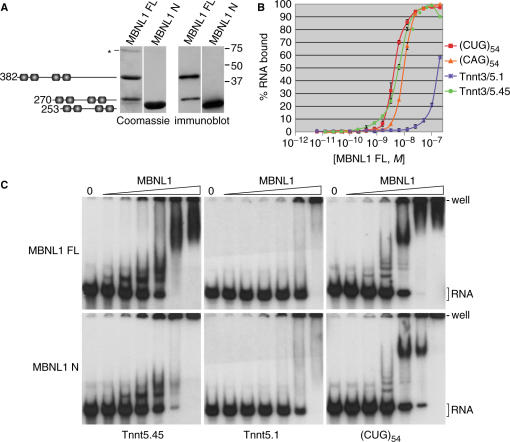
The MBNL1 proteins used for this study were either MBNL1 FL or the C-terminal truncation mutant MBNL1 N. For the MBNL1 FL preparation, ∼60% of the purified protein was full length while the MBNL1 N protein preparation was homogeneous as determined by Coomassie blue staining and immunoblot analysis (Figure 3A). Because MBNL1 shows a temperature-dependent binding profile in cell extracts in the absence of ATP (data not shown), all recombinant protein-binding studies were performed at 30°C to maximize binding while minimizing RNA degradation. As anticipated, MBNL1 FL showed a high affinity in the filter-binding assay for (CUG)54 (Kd = 5.3 ± 0.6 nM), but it also showed high affinities for Tnnt3 5.45 (Tnnt5.45) (Kd = 6.6 ± 0.5 nM) and (CAG)54 (Kd = 11.2 ± 1.5 nM) (Figure 3B). In contrast, Tnnt3/T5.1 RNA, which did not cross-link to MBNL1 (Figure 1B), bound poorly. The similarities in the binding affinities of MBNL1 for (CUG)54 and (CAG)54 accounts for the prior observation that overexpression of either of these repeat RNAs in COS-M6 cells results in the formation of nuclear foci that colocalize with GFP-MBNL1 (22). Relatively weak cooperativity was noted for interactions between MBNL1 and (CUG)54, (CAG)54 and Tnnt3 T5.45 RNAs (Hill coefficients of 1.62 ± 0.21, 1.67 ± 0.27 and 1.40 ± 0.14, respectively).
Figure 3.
MBNL1 binds to pathogenic and splicing precursor RNAs with similar affinities. (A) Purified recombinant MBNL1 proteins. Coomassie-stained gels (left panels) and immunoblots (right panels) of either full-length or N-terminal proteins. Illustration shows the primary structures (line; C3H motifs are shown as shaded boxes) indicating the full-length (382 aa) and N-terminal (253 aa) MBNL1 proteins together with a C-terminal truncated protein (270 aa) generated during expression in E. coli and an unknown 75 kDa protein (asterisk). (B) Nitrocellulose filter-binding analysis of MBNL1 FL binding to (CUG)54 (red square), (CAG)54 (orange triangle), Tnnt3 5.1(T5.1, blue cross) and Tnnt3 5.45 (T5.45, green circle) RNAs. (C) Gel shift analysis of MBNL1 FL and MBNL1 N binding to Tnnt3 5.45, 5.1 or (CUG)54. The positions of the free RNA (bracket), the gel origin (well) and MBNL1 FL and N concentrations (triangle, lanes are 0, 0.25, 1, 4, 16, 64, 256 nM) are indicated.
Comparable affinities were obtained when MBNL1–RNA complexes were analyzed by gel shift analysis (Figure 3C). Incubation of full-length MBNL1 (MBNL1 FL) with Tnnt3/T5.45 generated several protein–RNA complexes resolved by the polyacrylamide gel whereas significant binding to Tnnt3/T5.1 was only detectable at 256 nM and only gel excluded complexes were observed. Similar complexes were also formed with MBNL1 N although the truncated protein had a higher affinity for Tnnt3/T5.1 RNA and formed fewer gel-resolved complexes with Tnnt3/T5.45 and (CUG)54 (Figure 3C). Incubation of MBNL1 with (CUG)54 also resulted in the formation of several major complexes at protein concentrations (4–16 nM) near the Kd determined by filter binding. At higher MBNL1 FL concentrations (≥64 nM) the majority of the resulting MBNL1–(CUG)54 complexes migrated at, or near, the top of the gel. Similar gel shift profiles were obtained when RNA–protein complexes were formed in a higher salt buffer containing magnesium (see Methods section) although all Kd values increased ∼4-fold. The striking similarity in the binding affinities of MBNL1 for pathogenic and splicing precursor RNAs prompted us to re-examine the interaction of this splicing factor with CUGexp RNA.
MBNL1 binds to the stem region of a pathogenic dsRNA
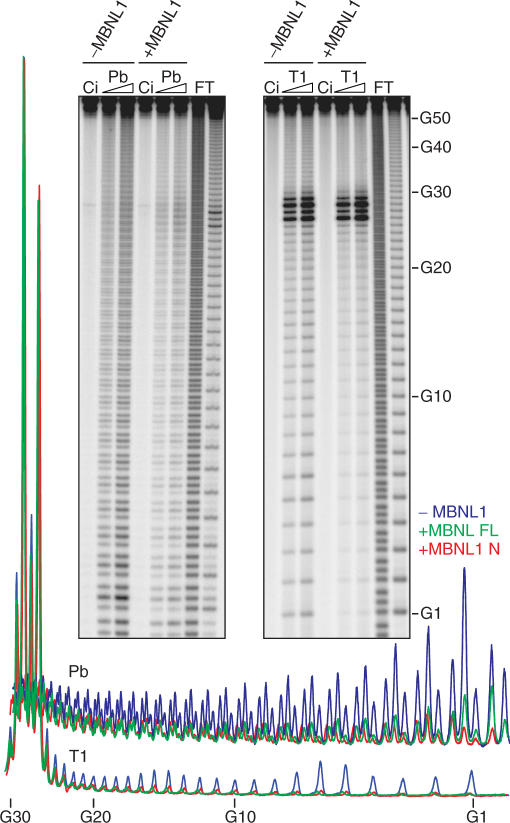
Muscleblind-like proteins were originally characterized as nuclear factors which are recruited by CUGexp RNAs (9). The predicted double-stranded nature of these pathogenic RNAs has been validated by chemical and nuclease mapping, thermal denaturation and electron microscopy (23–26). While the binding of MBNL proteins to CUGexp RNA is proportional to the predicted stem length, there is currently no direct experimental evidence that MBNL binds directly to the CUGexp stem or that the RNA structure remains in the hairpin configuration following MBNL binding. Therefore, we initially used chemical and enzymatic structure probing of labeled RNAs to identify MBNL1-binding sites on (CUG)54 RNA. RNAs were 5′ end-labeled, subjected to either lead ion (Pb)-induced hydrolysis or RNase T1 digestion in the absence or presence of recombinant MBNL1, and the products were fractionated on denaturing polyacrylamide gels. As shown previously, lead ions cleave both ssRNA and relaxed dsRNA structures which yielded a uniform ladder that increased in intensity with increasing lead concentration (Figure 4, left panel). As anticipated, addition of MBNL1 inhibited strand cleavage in a concentration-dependent manner and densitometry analysis failed to show significant regional differences in the cleavage pattern by MBNL1 suggesting uniform binding of this protein throughout the stem region. In contrast to lead, RNase T1 prefers to cleave after G nucleotides in single-stranded regions. Thus, incubation with RNase T1 resulted in strong cleavage at the terminal loop (Figure 4, right panel, G26–G29). Interestingly, terminal loop cleavage was unaffected by MBNL1 addition while stem cleavage was uniformly inhibited. We conclude that MBNL1 interacts primarily with the stem region of CUGexp RNAs.
Figure 4.
MBNL1 binds throughout the dsCUG stem. Structural analysis of 5′-end labeled (CUG)54 transcript (20 nM) either in the presence (+) or absence (−) of 500 nM recombinant MBNL1 and either lead ion (0.25 or 0.5 mM, lanes Pb) or ribonuclease T1 (0.5 or 1 U/µl, lanes T1); incubation control (no probe added, lane Ci); formamide (lane F); guanine-specific ladder obtained with RNase T1 digestion (lane T). The positions of selected G residues are shown along the T1 ladder (G-residues of the corresponding CUG repeat are indicated). Below the gel panels is the color-coded densitometric analysis (ImageQuant) of cleavage patterns for the CUG stem obtained with lead ion (Pb) and T1 ribonuclease (T1) in the presence of MBNL1 FL (green), MBNL1 N (red) or in the absence of protein (blue).
Visualization of MBNL1–CUGexp complexes
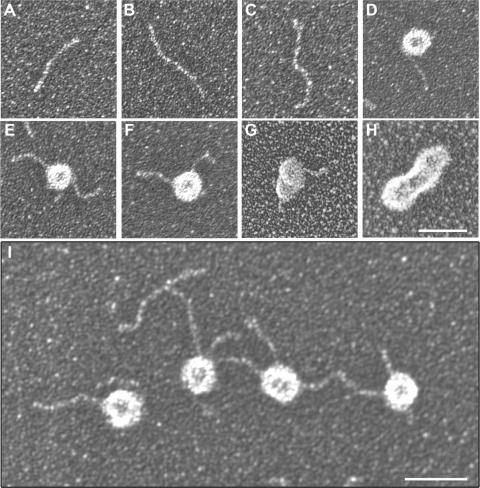
In a previous study, we reported that the MBNL splicing antagonist, CUGBP1, binds to out-of-register ssCUG repeats at the base of CUGexp RNA hairpins but not the A-form helical region while the dsRBD protein TRBP associates with the stem region (25). To confirm that MBNL1 is a stem-binding protein, electron microscopy (EM) was performed using the full-length MBNL1 protein. Purified (CUG)136 was examined following direct absorption to thin carbon foils, dehydration and tungsten shadowing. In contrast to ssRNA, (CUG)136 RNA formed rod-like segments as described previously for (CUG)130 (Figure 5A–C). To examine the structures of MBNL1–RNA complexes, MBNL1 was incubated with RNAs at two different RNA:protein molar ratios (1:2.5 or 1:10) and subsequently prepared for EM. In the presence of RNA, purified MBNL1 formed a ring-shaped structure with a prominent central cavity and for (CUG)136–MBNL1 complexes incubated at a ratio of 1:2.5, ∼70% of the RNAs were bound by one of these MBNL1 rings (Figure 5D–F). At higher protein levels (1:10), free (CUG)136 RNA was rarely detectable (6.4% of the RNAs in the field) while >90% of the RNAs were bound by two or more MBNL1 rings (Figure 5G and H). Also shown is a representative field of dsCUG RNAs and MBNL1–(CUG)136 complexes (Figure 5I, RNA:protein is 1:2.5). Although the majority of MBNL1 rings were associated with RNA under our RNA assembly conditions, a few free rings were visualized in the background in the absence of associated (CUG)136 helices suggesting that MBNL1 may form a ring structure independent of RNA. We failed to visualize rings by negative staining suggesting that this structure is disrupted by the acidic conditions of the negative staining protocol. Interestingly, when MBNL1 N proteins were incubated with (CUG)136, ring structures were observed although ring size was much less uniform and there was considerably less stacking of MBNL1 N rings compared to MBNL1 FL (data not shown). This observation prompted us to examine whether the C-terminal region was important for MBNL1 homotypic interactions.
Figure 5.
Visualization of dsCUG and MBNL1–dsCUG complexes. Electron microscopy of either free (CUG)136 RNAs (A–C) or MBNL1–(CUG)136 complexes (D–I) at a protein:RNA ratio of either 2.5:1 (D–F, I) or 10:1 (G–H). As reported previously, purified dsCUG RNAs, which were directly adsorbed onto thin carbon foils followed by dehydration and rotary shadow casting with tungsten, are elongated rod-shaped structures (25). For analysis of complexes, MBNL1 protein and (CUG)136 RNA were incubated together at 30°C, fixed with glutaraldehyde, passed over a gel filtration column and adsorbed on copper grids for rotary shadowing. Under these conditions, MBNL1 has a distinct ring-shaped structure. Size bars (white line) are 40 nm (H) or 50 nM (I).
MBNL1 self-interaction mediated by the C-terminal region
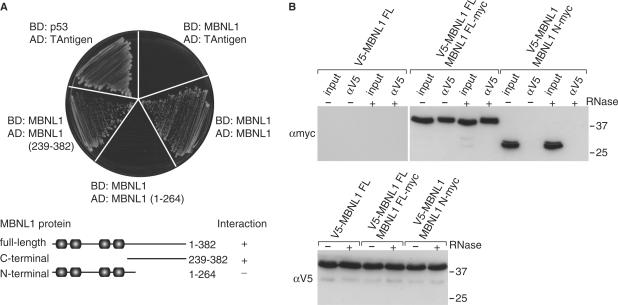
A number of studies have demonstrated that MBNL1 accumulates in ribonuclear foci together with pathogenic RNAs (13,27–31). While additional proteins might also bind to CUGexp and CCUGexp RNAs, the number of ribonuclear foci in DM myoblasts declines significantly following loss of MBNL1 suggesting that this protein is required for the formation and/or maintenance of these unusual nuclear structures (15). Since many RNA-binding proteins function as components of large multi-subunit complexes (32) and some of these proteins self-interact via their auxiliary or non-RNA-binding regions (33), we tested the possibility that MBNL1 proteins self-associate using the yeast two-hybrid system. Although the amino terminal region of MBNL1 contains all four C3H motifs and is responsible for protein–RNA interactions (21), very little is known about the function of the C-terminal region. The MBNL1 FL protein showed strong homotypic interactions in this system (Figure 6A). Although MBNL FL failed to interact with the MBNL1 N-terminal region (1–264), interactions between the full-length protein and C-terminal region (239–382) were readily detectable. This C-terminal region does not contain any known RNA binding, or other, structural motifs.
Figure 6.
Self-association of MBNL1 proteins is mediated by the C-terminal region. (A) Two-hybrid analysis using yeast strains transformed with the following binding domain (BD) and activation domain (AD) plasmids: (i) GAL4 DNA-binding domain (BD) plasmids with either p53 (activation control) or full-length MBNL1(MBNL1); (ii) activation domain plasmids with either T-antigen (activation control), MBNL1 (residues 1–382), MBNL1 1–264 (N-terminal region) or MBNL 239–382 (C-terminal region). Functional interactions between the proteins expressed from the BD and AD plasmids results in growth on the Trp− Leu− His− selection plate. (B) Co-immunopurification of MBNL1 requires the C-terminal region. HEK293T cells were co-transfected for 24 h with plasmids expressing tagged versions of either full-length or N-terminal MBNL1 (V5-MBNL1 FL alone, co-transfected V5-MBNL1 FL and MBNL1 FL-myc, co-transfected V5-MBNL1 FL and MBNL1 N-myc). Cell lysates were prepared and the V5-MBNL1 FL protein immunopurified (αV5) followed by SDS–PAGE (50% of IP sample) and immunodetection of MBNL1 FL-myc or MBNL1 N-myc using mAb 9E10 (top panel, input lanes represent 2.5% of total IP) or V5-MBNL1 FL (bottom panel, only the input lanes are shown).
To confirm that MBNL1 homotypic interactions occurred in a mammalian cell context, 293T cells were co-transfected with plasmids which expressed either V5-MBNL1 FL alone, V5-MBNL1 FL and MBNL1 FL-myc or V5-MBNL1 FL and MBNL1 N-myc. Twenty-four hours following transfection, V5-tagged MBNL1 was immunopurified from cell lysates using an anti-V5 antibody and the precipitates were then immunoblotted using either anti-myc or anti-V5 antibodies. In agreement with the two-hybrid analysis, the full-length V5 and myc-tagged proteins were associated in vivo while the N-terminal MBNL1 region (MBNL1 N-myc) failed to co-immunopurify with V5-MBNL1 FL (Figure 6B). Interactions between V5-MBNL1 FL and MBNL1 N-myc were not mediated by RNA tethering since treatment of the cell lysate with RNase A did not affect the amount of MBNL1 N-myc in the V5-MBNL1 FL immunoprecipitate. Although it is possible that MBNL1 interactions could be mediated by other nuclear factors, our demonstration that purified MBNL1 forms a ring-like structure argues against this interpretation. We conclude that MBNL1–MBNL1 interactions occur in vivo and these interactions are mediated by the C-terminal region.
DISCUSSION
MBNL1 targets similar binding motifs in splicing precursor and pathogenic RNAs
A number of studies have demonstrated that CUGexp and CCUGexp RNAs sequester MBNL proteins in nuclear RNA foci of DM cells (9,13,15,27–31,34,35). Loss of MBNL1 function by sequestration correlates with the inhibition of alternative splicing regulation for a specific set of developmentally regulated exons that are mis-spliced in DM tissues (8,15,20,36). Moreover, Mbnl1 knockout mice develop many of the characteristic features associated with DM disease, including myotonia and subcapsular cataracts, while adeno-associated virus (AAV)-mediated Mbnl1 overexpression in transgenic mice expressing a CUGexp in skeletal muscle reverses myotonia and DM-associated mis-splicing (16,20).
Although there is considerable evidence for this muscleblind loss-of-function model for DM pathogenesis, the molecular basis for MBNL1 sequestration by CUGexp and CCUGexp RNAs has not been elucidated. Since mutant RNA expansions must compete with normal pre-mRNA, and possibly mRNA, binding sites for MBNL1 recruitment, effective MBNL1 sequestration might occur if the affinity of this protein for CUGexp and CCUGexp RNAs is greater than for its normal splicing targets. The binding analysis presented here does not support this conjecture since MBNL1 also possesses relatively high affinities for CAGexp and Tnnt3 precursor RNAs. Indeed, these binding studies provide an explanation for the formation of MBNL1-containing ribonuclear foci in cells overexpressing CAGexp (22). Additionally, mapping of a binding site to a stem-loop structure in Tnnt3 intron 8 just upstream of the F exon indicates that RNA recognition by MBNL proteins involves a common interaction mode for both pathogenic and normal pre-mRNAs: recognition of GC-rich hairpins containing pyrimidine mismatches.
What is the physiological significance of the binding preference of MBNL1 for RNA stem-loop structures? In this study, we provide evidence that MBNL1 is a developmentally regulated splicing regulator which acts as an intronic splicing repressor by recognizing a stem-loop near the Tnnt3 F exon 3′ splice site, possibly resulting in the stabilization of this secondary structure and interference with U2AF recruitment. Based on this observation, we speculate that MBNL1 recognizes similar intronic stem-loop structures adjacent to the 5′ splice sites of other MBNL1-regulated fetal exons to inhibit U1 snRNP recruitment or MBNL1 may stabilize interactions between introns flanking alternative exons resulting in RNA looping and increased fetal exon skipping.
The importance of RNA secondary structures in inherited disease and alternative splicing has been previously highlighted in frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) which is caused by mutations in the MAPT gene encoding the microtubule-associated protein tau (37). Some FTDP-17 mutations destabilize a predicted stem-loop structure, which forms between the 3′end of exon 10 and the 5′ end of the downstream intron, resulting in an increase in U1 snRNP recruitment and E10 inclusion. Interestingly, MAPT exon 10 skipping increases in the DM brain suggesting that MBNL1 promotes exon 10 inclusion during splicing (20). The MBNL1-binding preferences shown in this study suggest that this factor may also function as a splicing activator by recognizing RNA stem-loop structures in novel exonic and/or intronic splicing enhancers or by blocking splicing silencer elements by stabilizing overlapping RNA secondary structures.
Another interesting question arose when we mapped the MBNL1-binding site on Tnnt3 to an 18-nt stem-loop structure. Our previous study showed that MBNL1 in HeLa nuclear extract cross-links strongly to (CUG)74 and (CUG)97 but not to (CUG)<20 (9). Based on this observation, we proposed that below a certain length threshold (<20 repeats) the dsCUG helix was unstable in the cell extract and ssCUG was not a binding site for MBNL1. The results reported here support that proposal and demonstrate that MBNL1 is primarily a dsRNA-binding protein which recognizes relatively short GC-rich hairpins if the overall RNA secondary structure is stabilized by additional sequence interactions. This conclusion provides a plausible resolution for the apparently conflicting results that overexpression of a GFP-DMPK 3′-UTR (CTG)5 transgene results in a DM phenotype (14) while mice expressing the HSASR (human skeletal α-actin containing a 3′-UTR with five CTG repeats) transgene are normal (38). For the DMPK 3′-UTR, the (CTG)5 repeat is predicted to interact with an upstream region to form a GC-rich stem interrupted by several U-U and C-C mismatches while this repeat in the HSA 3′-UTR is located in sequential unpaired loops. We postulate that this structural arrangement in the DMPK 3′-UTR promotes MBNL1 sequestration when GFP-DMPK 3′-UTR (CTG)5 RNA is overexpressed in transgenic mice.
MBNL1 rings: interactions with CUGexp RNA and potential relevance to DM disease
Filter binding and gel shift assays indicated that the affinity of MBNL1 for Tnnt3/T5.45 lies between that of (CUG)54 and (CAG)54. While overexpression of either CUG or CAG repeats induces the formation of nuclear foci in cell culture, it is interesting to note that we did not observe RNA foci formation following Tnnt3 minigene overexpression. This observation argues that high affinity MBNL1–RNA interactions together with abundant expression of MBNL1 target RNAs is not sufficient for ribonuclear foci formation. Of course, MBNL1 may be cleared from splicing target, but not pathogenic, RNAs during RNA processing and nuclear export or unusual interactions between MBNL proteins and CUGexp RNA might drive foci formation. In support of the latter possibility, we provide EM evidence that MBNL1 forms a tandem ring structure when bound to CUGexp RNA. The size of the rings is uniform with a diameter of ∼18 nm and since the MBNL1 isoform employed for these studies is 41 kDa, the ring structure must be an oligomeric complex. At a protein:RNA ratio of 2.5:1, most CUGexp helices were bound by a single ring while the majority of these hairpins were bound by at least two rings at a higher protein:RNA ratio. It is not clear if the MBNL1 ring contains a hole but if a central cavity exists it might allow threading of dsRNA. When multiple rings were bound to a single RNA molecule they appeared to be tandemly stacked suggesting either a preferential ring-loading site or potential ring–ring interactions. The latter possibility is supported by our finding that MBNL1 self-interacts via its C-terminal region both in the yeast two-hybrid system and in mammalian cells. In contrast, the MBNL1 N-terminal region encompassing the CCCH RNA-binding motifs fails to interact with full-length MBNL1 although RNA-binding activity is comparable to the full-length MBNL1 (data not shown). In this regard, it is interesting to note that the rings formed using MBNL1 N, lacking the C-terminal region, were less uniform in size and there was less ring–ring stacking at higher protein:RNA ratios. A similar situation has been noted for the Escherichia coli protein Hfq (Host factor 1) which is a single-strand RNA-binding protein involved in the translational regulation and stability of several RNAs (39). As visualized in the EM by negative staining, Hfq forms hexameric rings. Similar to MBNL1, the RNA-binding activity of Hfq resides in the N-terminal region and rings are still formed by a C-terminal truncated protein although they are less stable.
Does MBNL1 exist as a ring structure when bound to its normal RNA splicing targets? To address this question, we performed EM analysis of the MBNL1–Tnnt3/T5.45 complex. Although MBNL1 rings were observed, the result was inconclusive due to the difficulty in visualizing small and partially single-stranded Tnnt3/T5.45 RNA by EM. However, MBNL1 forms large complexes with both CUG and Tnnt3/T5.45 RNAs (Figure 3C) so it is possible that MBNL1 forms a ring structure when bound to splicing regulatory sites. Nevertheless, these normal binding targets do not contain the tandemly arrayed MBNL1-binding sites present on pathogenic CUGexp RNAs which we propose are essential for multiple ring interactions, MBNL1 sequestration and ribonuclear foci formation in DM cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Keith Nykamp for help with the yeast two-hybrid analysis, Rob Osborne for the gift of pBC-CTG136, members of the Swanson lab for comments on the manuscript and Andy Berglund for communicating results prior to publication. This work was supported by grants to M.S.S. from the NIH and the Paul D. Wellstone Muscular Dystrophy Cooperative Research Center (AR46799, NS48843) and a post-doctoral fellowship to K.S. from the Foundation for Polish Science. The electron microscopy studies were supported by NIH grants GM31819 and ES013773 to J.D.G. Funding to pay the Open Access publication charges for this article was provided by NIH AR46799.
Conflict of interest statement. None declared.
REFERENCES
- 1.Perez-Canadillas JM, Varani G. Recent advances in RNA-protein recognition. Curr. Opin. Struct. Biol. 2001;11:53–58. doi: 10.1016/s0959-440x(00)00164-0. [DOI] [PubMed] [Google Scholar]
- 2.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 3.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 4.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 5.Chang KY, Ramos A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005;272:2109–2117. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- 6.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu D, Searles MA, Klug A. Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature. 2003;426:96–100. doi: 10.1038/nature02088. [DOI] [PubMed] [Google Scholar]
- 8.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nykamp KR, Swanson MS. Toxic RNA in the nucleus: unstable microsatellite expression in neuromuscular disease. Prog. Mol. Subcell. Biol. 2004;35:57–77. doi: 10.1007/978-3-540-74266-1_3. [DOI] [PubMed] [Google Scholar]
- 11.Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 12.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 13.Mankodi A, Teng-Umnuay P, Krym M, Henderson D, Swanson M, Thornton CA. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann. Neurol. 2003;54:760–768. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- 14.Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenzel-McCardell CD, Bourne TD, Phillips LH. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- 16.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 17.Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 18.Soller M, White K. ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev. 2003;17:2526–2538. doi: 10.1101/gad.1106703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reichert V, Moore MJ. Better conditions for mammalian in vitro splicing provided by acetate and glutamate as potassium counterions. Nucleic Acids Res. 2000;28:416–423. doi: 10.1093/nar/28.2.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 22.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 23.Napierala M, Krzyzosiak WJ. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 24.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michalowski S, Miller JW, Urbinati CR, Paliouras M, Swanson MS, Griffith J. Visualization of double-stranded RNAs from the myotonic dystrophy protein kinase gene and interactions with CUG-binding protein. Nucleic Acids Res. 1999;27:3534–3542. doi: 10.1093/nar/27.17.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 28.Mankodi A, Lin X, Blaxall BC, Swanson MS, Thornton CA. Nuclear RNA foci in the heart in myotonic dystrophy. Circulation Res. 2005;97:1152–1155. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- 29.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 30.Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 32.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 33.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 34.Schoser BG, Kress W, Walter MC, Halliger-Keller B, Lochmuller H, Ricker K. Homozygosity for CCTG mutation in myotonic dystrophy type 2. Brain. 2004;127:1868–1877. doi: 10.1093/brain/awh210. [DOI] [PubMed] [Google Scholar]
- 35.Wheeler TM, Krym MC, Thornton CA. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul. Disord. 2007;17:242–247. doi: 10.1016/j.nmd.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent postnatal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat. Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 38.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 39.Arluison V, Folichon M, Marco S, Derreumaux P, Pellegrini O, Seguin J, Hajnsdorf E, Regnier P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004;271:1258–1265. doi: 10.1111/j.1432-1033.2004.04026.x. [DOI] [PubMed] [Google Scholar]