Abstract
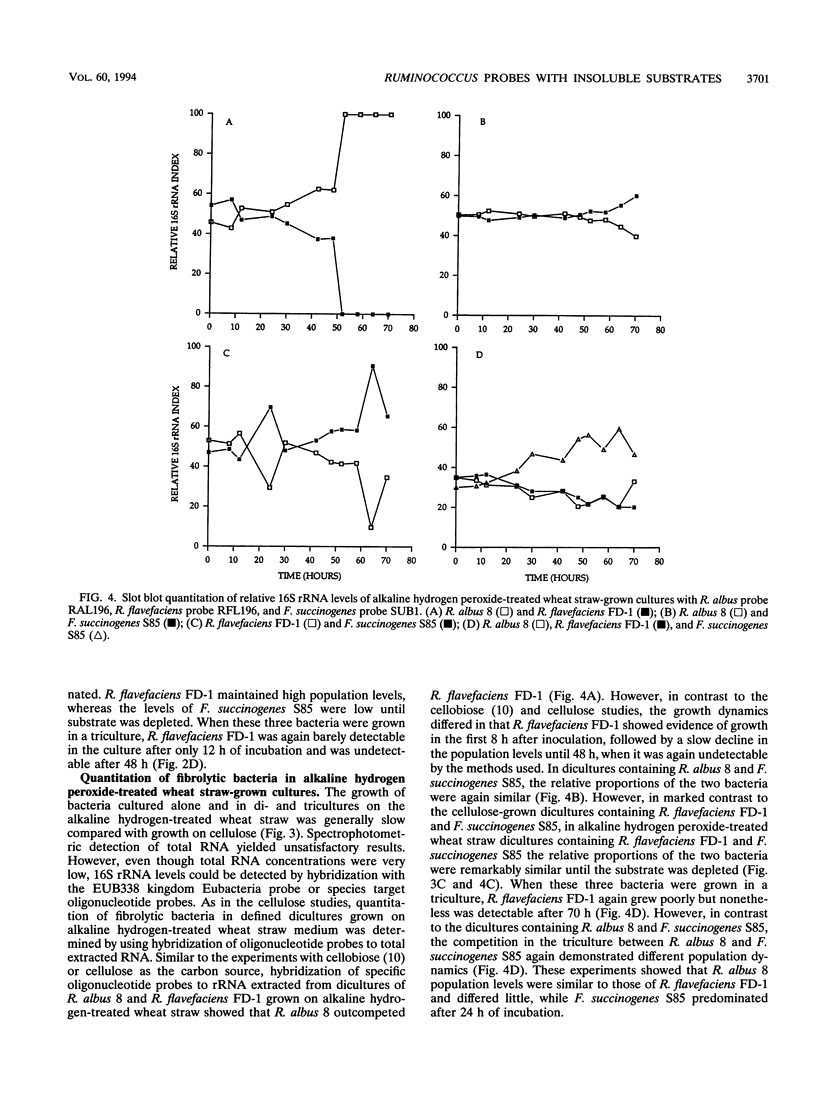
Specific oligonucleotide probes targeted to sites on the 16S rRNA of Ruminococcus albus 8, Ruminococcus flavefaciens FD-1, and Fibrobacter succinogenes S85 and a domain Bacteria probe were used to study bacterial interactions during the fermentation of cellulose and alkaline hydrogen peroxide-treated wheat straw in monocultures, dicultures, and tricultures. Results showed that R. albus 8 inhibited the growth of R. flavefaciens FD-1 when grown as a diculture with cellulose or alkaline hydrogen peroxide-treated wheat straw as the carbon source. In dicultures containing R. albus 8 and F. succinogenes S85 grown on cellulose or alkaline hydrogen peroxide-treated wheat straw, competition was not detected. R. flavefaciens FD-1 outcompeted F. succinogenes S85 when cellulose was used as the carbon source. In tricultures with cellulose as the carbon source, R. flavefaciens FD-1 was inhibited, R. albus 8 appeared to dominate during the early phase of degradation (12 to 48 h), while F. succinogenes S85 became predominant during the later phase of degradation (60 to 70 h). When alkaline hydrogen peroxide-treated wheat straw was used as a growth substrate, F. succinogenes S85 showed better growth than either R. albus 8 or R. flavefaciens FD-1. However, R. flavefaciens FD-1 was present in small numbers throughout the incubation period, unlike the growth patterns when cellulose was the carbon source.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Krumholz L., Stahl D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990 Feb;172(2):762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Hahn D., Starrenburg M. J., Akkermans A. D. Oligonucleotide Probes That Hybridize with rRNA as a Tool To Study Frankia Strains in Root Nodules. Appl Environ Microbiol. 1990 May;56(5):1342–1346. doi: 10.1128/aem.56.5.1342-1346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenyo A. A., Mackie R. I., Fahey G. C., Jr, White B. A. Degradation of wheat straw and alkaline hydrogen peroxide-treated wheat straw by Ruminococcus albus 8 and Ruminococcus flavefaciens FD-1. J Anim Sci. 1991 Feb;69(2):819–826. doi: 10.2527/1991.692819x. [DOI] [PubMed] [Google Scholar]
- Odenyo A. A., Mackie R. I., Stahl D. A., White B. A. The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: development of probes for Ruminococcus species and evidence for bacteriocin production. Appl Environ Microbiol. 1994 Oct;60(10):3688–3696. doi: 10.1128/aem.60.10.3688-3696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]


