Abstract
In several forms of β-thalassemia, mutations in the second intron of the β-globin gene create aberrant 5′ splice sites and activate a common cryptic 3′ splice site upstream. As a result, the thalassemic β-globin pre-mRNAs are spliced almost exclusively via the aberrant splice sites leading to a deficiency of correctly spliced β-globin mRNA and, consequently, β-globin. We have designed a series of vectors that express modified U7 snRNAs containing sequences antisense to either the aberrant 5′ or 3′ splice sites in the IVS2–705 thalassemic pre-mRNA. Transient expression of modified U7 snRNAs in a HeLa cell line stably expressing the IVS2–705 β-globin gene restored up to 65% of correct splicing in a sequence-specific and dose-dependent manner. Cell lines that stably coexpressed IVS2–705 pre-mRNA and appropriately modified U7 snRNA exhibited up to 55% of permanent restoration of correct splicing and expression of full-length β-globin protein. This novel approach provides a potential alternative to gene replacement therapies.
Keywords: gene therapy, antisense, genetic disease, thalassemia
Gene therapy appears to be the most promising treatment for genetic disorders (reviewed in ref. 1). It is usually understood as either the replacement of a defective gene with the correct one or expression of a transgene whose product supplants its defective counterpart. These forms of gene therapy have been tested in animal models and in the clinic, for example, in treatment of adenosine deaminase deficiency (1, 2), cystic fibrosis (3), and other genetic disorders (4, 5). Although, in principle, gene therapy should be applicable to any gene-based disorder, the difficulties with vectors suitable for efficient delivery of large transgenes or providing sustained expression of the transfected genes in a tissue-specific, properly regulated manner (6, 7) limit its clinical applicability. Regulated expression is especially important in gene therapy for correction of tightly regulated genes such as β-globin in sickle cell anemia or thalassemia. Expression of the β-globin transgene is useful only if it occurs in concert with the α-globin genes in erythroid cells. Although the β-globin gene is small, its regulated expression is difficult to achieve because it is controlled by a large locus control region. Vectors capable of accommodating large fragments of DNA are not yet available, and truncated constructs, in spite of significant progress, do not provide the desired levels and specificity of expression (8–10).
In addition to gene replacement, gene therapy may also be accomplished by manipulation of gene structure and expression. It has been shown recently in model cell culture systems that double-stranded chimeric RNA–DNA oligonucleotides may induce site-specific removal from the human β-globin gene of the mutation responsible for sickle cell anemia (11). Clinically relevant alteration of globin gene expression also can be achieved by relatively simple pharmacological treatments. For example, hydroxyurea or butyric acid and its derivatives induce the expression of fetal hemoglobin, which partially compensates for the lack of correct β-globin expression in sickle cell anemia or thalassemia. These treatments were successful in clinical trials (12–15).
Work in this laboratory showed that antisense oligonucleotides may restore the activity of thalassemic β-globin genes carrying mutations that cause defects in pre-mRNA splicing. Oligonucleotides targeted to the aberrant splice sites generated by the thalassemic mutations in intron 2 of the β-globin gene—IVS2–654, -705, and -745 (refs. 16 and 17; unpublished data)—blocked the aberrant splice sites and restored the correct splicing pattern by forcing the splicing machinery to reselect the existing correct splice sites. The correction of splicing was accompanied by translation of the resultant β-globin mRNA into full-length β-globin protein. If the same results were achieved in the erythroblasts of a thalassemic patient, a more balanced synthesis of α- and β-globin would be restored and the clinical symptoms of thalassemia would be ameliorated. Note that in patients, the antisense oligonucleotides would have restored correct splicing of pre-mRNA, properly transcribed from the β-globin gene in its natural chromosomal environment. This precludes the possibility of overexpression of β-globin mRNA, an important consideration in the treatment of hemoglobinopathies. However, a significant drawback of this approach stems from the fact that the oligonucleotides do not remove the mutation and therefore would require periodic administrations.
To circumvent this problem we have developed an approach that allows for long-term, possibly permanent, expression of RNA antisense to aberrant thalassemic splice sites in β-globin pre-mRNA. This was accomplished by incorporating the anti-β-globin sequences into the gene for murine U7 small nuclear RNA (snRNA). U7 snRNA, in a complex with at least two U7-specific proteins and eight common Sm proteins (18), forms a ribonucleoprotein particle (U7 snRNP) that is involved in the processing of the 3′ end of histone pre-mRNAs (19–21). We show here that the insertion of appropriate antisense sequences into the U7 snRNA changed its function from a mediator of histone mRNA processing to an effector of alternative splicing of β-globin pre-mRNA. Stable transfection of cells expressing thalassemic β-globin gene with vectors carrying a modified U7 snRNA gene led to a permanent correction of the splicing pattern of the β-globin pre-mRNA. This resulted in the accumulation of significant amounts of full-length β-globin mRNA and the corresponding protein.
MATERIALS AND METHODS
U7 snRNA Constructs.
The U7SmOPT plasmid carries the mouse U7 snRNA gene in which the U7-specific Sm binding site (AAUUUGUCUAG) was replaced with the consensus Sm sequence (AAUUUUUGGAG) (22). The U7 promoter and 3′ sequences are included in the construct. In U7.3, U7.5, U7.34, and U7.324 constructs, the natural 18-nt sequence complementary to the 3′ processing site of histone pre-mRNAs was replaced (23, 24) with sequences complementary to either the 3′ or the 5′ splice sites activated by the IVS2–705 mutation (see Fig. 1C). The details of the construction are available on request.
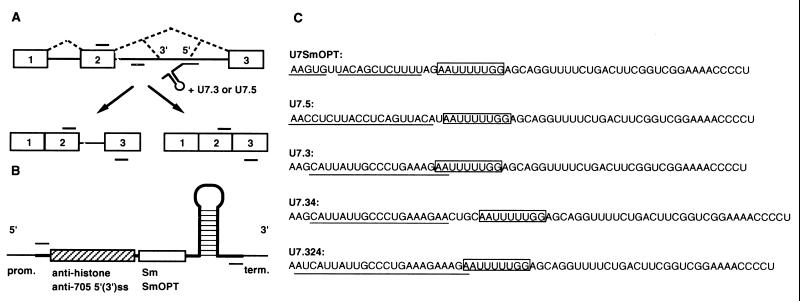
Figure 1.
(A) Correction of splicing of IVS2–705 β-globin pre-mRNA by modified U7 snRNA. Boxes, exons; lines, introns; short bars above and below RNA, primers used in PCR and RT-PCR analysis. The dashed lines represent correct and aberrant splicing pathways. The modified U7 snRNA targeted to the IVS2–705 splice site (5′) is depicted under the pre-mRNA. (B) Structure of U7 snRNA constructs. Wild-type U7 snRNA (heavy line) includes a stem-loop structure, the U7-specific Sm sequence (open box), and a sequence antisense to the 3′ end of histone pre-mRNA (stippled box). In anti-705 U7 snRNAs, the two sequences are replaced with the SmOPT sequence and with antisense sequences to the aberrant 3′ or 5′ splice sites in the β-globin gene, respectively. The promoter (prom.) and 3′ end-forming (term.) regions are indicated. Short bars above and below the U7 construct represent primers used in PCR and RT-PCR analysis. (C) Sequences of U7 snRNA constructs. The Sm-binding site is boxed and the antisense sequences are underlined.
Cell Lines.
The HeLa cell line carrying the thalassemic IVS2–705 human β-globin gene (25) and the cell lines stably expressing the modified U7 snRNAs were grown in MEM with 5% fetal calf and 5% horse sera. The latter cell lines were obtained by cotransfection of the HeLa IVS2–705 cells with a plasmid carrying a hygromycin-resistance gene and a U7 snRNA-expressing plasmid (see legend to Fig. 6) in the presence of Lipofectamine (8 μg/ml, Life Technologies) as recommended by the manufacturer. Stable transfectants were isolated after selection in medium containing 250 μg/ml hygromycin.
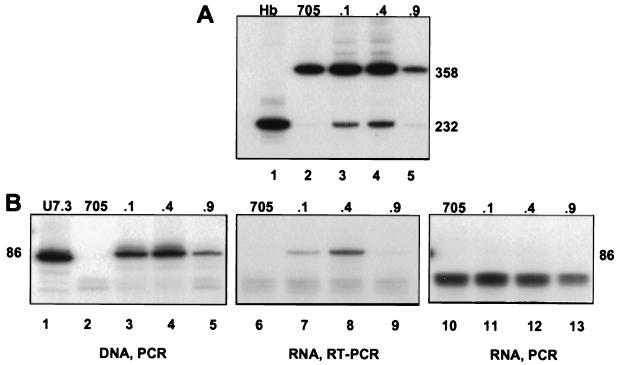
Figure 6.
(A) Correction of splicing in cell lines stably expressing U7.324 snRNA. The IVS2–705 cell line was cotransfected with the U7.324 construct and a plasmid carrying the hygromycin-resistance gene. Total RNA from the resulting hygromycin-resistant cell lines was analyzed by RT-PCR. Lanes: 1, RNA from human blood; 2, RNA from IVS2–705 cell line; 3–5, stable correction of splicing in three independent cell lines (705U7.324.1, .4, and .9) as indicated at the top of the figure. (B) Correlation of splicing correction with stable expression of U7.324 snRNA. U7.3 plasmid (200 ng, lane 1), total DNA (lanes 2–5), or total RNA (lanes 6–13) from the cell lines shown in Fig. 6A were analyzed by PCR and RT-PCR, respectively, by using U7-specific primers (see Fig. 1B and Materials and Methods). Because isolated RNA possibly could be contaminated with genomic DNA, in lanes 10–13 the RNA was analyzed with the reverse transcription step omitted in the assay.
Transient Expression of Modified U7 snRNA.
For all experiments, HeLa IVS2–705 cells were plated 24 hr before treatment in 24-well plates at 105 cells per 2-cm2 well. The cells were treated for 10 hr with modified U7 plasmids (0.5, 1, and 2 μg/ml) complexed with 8 μg/ml of Lipofectamine. Unless otherwise indicated, the cellular RNA or protein was isolated 24 hr after the end of transfection.
RNA and DNA Analysis.
Total cellular RNA or DNA was isolated by using TRI-Reagent (MRC, Cincinnati). RNA (200 ng) was analyzed by reverse transcription–PCR (RT-PCR) using rTth DNA polymerase as directed by the manufacturer (Perkin–Elmer). To maintain the linear concentration-dependent response, the PCR was carried out for 18 cycles (26) with the addition of 0.2 μCi of [α-32P]dATP (1 Ci = 37 GBq) to the PCR mixture. Correction of human β-globin pre-mRNA splicing was detected with forward and reverse primers spanning positions 21–43 of exon 2 and positions 6–28 of exon 3, respectively, in β-globin mRNA. Quantitation of β-globin pre-mRNA was performed with forward and reverse primers spanning positions 21–43 of exon 2 and positions 119–142 of intron 2. Expression of modified U7 snRNA was assayed with forward (GCATAAGCTTAAGCATTATTGCCCTGAA) and reverse (CGTAGAATTCAGGGGTTTTCCGACCGA) primers; underlined nucleotides overlap with U7 sequences. RT-PCR products were separated on 7.5% nondenaturing polyacrylamide gels. The gels were dried and autoradiographed with Kodak BioMax film. For the control experiment shown in Fig. 6B, 200 ng of chromosomal DNA was subjected to PCR with the same U7 specific primers.
Protein Analysis.
Hemin (10 μM, Fluka) treatment of transfected cells was in serum-free medium for 4 hr immediately preceding the isolation of protein. Blots of proteins separated on a 10% Tricine-SDS polyacrylamide gel (27) were incubated with polyclonal affinity-purified chicken anti-human hemoglobin IgG as primary antibody and rabbit anti-chicken horseradish peroxidase-conjugated IgG as secondary antibody (Accurate Chemicals). The blots were developed with an enhanced chemiluminescence detection system (Amersham).
Image Processing.
All autoradiograms were captured by a Dage–MTI CCD72 video camera (Michigan City, IN), and the images were processed using nih image 1.57 and macdraw pro 1.0 software. The final figures were printed on a Tektronix Phaser 550 printer. nih image 1.57 also was used for quantitation of the autoradiograms. Correctly spliced β-globin mRNA was quantified by densitometry of the autoradiograms, with the results expressed as the percentage of correct product relative to the sum of the correct and aberrant products. The results were corrected to account for the higher [32P]dAp content of the PCR product derived from aberrantly spliced mRNA than that from correctly spliced mRNA.
RESULTS
In the thalassemic IVS2–705 human β-globin gene, a T-to-G mutation at position 705 of intron 2 improves the match of the surrounding sequence to the consensus donor (5′) splice site (ACTGAT/GTAAGA to ACTGAG/GTAAGA; slash indicates the splice site). In the transcribed IVS2–705 pre-mRNA, the presence of this new 5′ splice site activates an acceptor (3′) splice site 126 nt upstream, resulting in incorrectly spliced β-globin mRNA containing a fragment of the intron (Fig. 1A). This fragment creates a premature stop codon resulting in a truncated β-globin polypeptide. Thus, in individuals homozygous for this mutation, the levels of the β-globin subunit of hemoglobin are drastically reduced, leading to β-thalassemia (28).
To improve the method of correction of splicing by antisense oligonucleotides (see Introduction), we have introduced sequences encoding fragments antisense to the aberrant splice sites into the U7 snRNA gene and used these constructs to transfect cells expressing the IVS2–705 pre-mRNA. It was anticipated that this approach would result in long-term expression of antisense RNA. The choice of U7 snRNA and the design of the constructs (Fig. 1B) as antisense carriers was based on several considerations.
The first 18 nt of this 62-nt-long RNA function as a natural antisense sequence by hybridizing with the so-called spacer element of histone pre-mRNA during its 3′ processing (29, 30). Thus, it seemed likely that upon replacement of the anti-histone sequence with a sequence complementary to aberrant splice sites in IVS2–705 pre-mRNA, the resulting U7 snRNA molecule would bind equally well to the new target sequences and correct aberrant splicing in a manner similar to antisense oligonucleotides.
Endogenous U7 snRNA is expressed at a low level, approximately 2–15 × 103 molecules per cell. However, it was found that the expression level and the nuclear concentration of U7 snRNA could be increased significantly by converting the wild-type U7 Sm-binding site (AAUUUGUCUAG) to the consensus Sm-binding sequence derived from the major spliceosomal snRNPs (SmOPT, AAUUUUUGGAG) (31). Moreover, the SmOPT modification of U7 snRNA rendered the particle functionally inactive in histone pre-mRNA processing (22, 31). This potentially has two beneficial effects: (i) the target RNA, such as β-globin pre-mRNA, will not be cleaved by the histone 3′ end processing machinery, and (ii) because of the inability of U7 SmOPT particles to bind one or more U7-specific proteins (22), the RNA will not compete with endogenous U7 snRNP for potentially limiting U7-specific proteins. Finally, whereas the wild-type U7 snRNPs are sequestered in coiled bodies, those with the SmOPT modification are not (32) and therefore may be redirected to the sites of pre-mRNA splicing. Thus, the U7 gene with the SmOPT sequence was used to construct vectors expressing anti-705 U7 snRNAs (Fig. 1 B and C) with the assumption that the increased nuclear concentration of the RNA and the lack of competition from the wild-type molecule would improve its ability to block aberrant splice sites in IVS2–705 pre-mRNA.
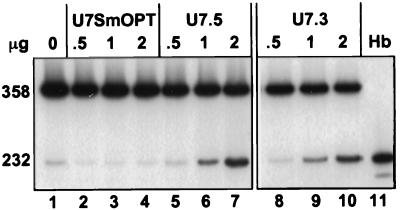
Fig. 2 shows the results of RT-PCR analysis of total RNA isolated 24 hr after transient transfection of a HeLa cell line expressing thalassemic IVS2–705 pre-mRNA with U7 constructs targeted to either of the aberrant splice sites. Both the U7 snRNA targeted to the aberrant 5′ splice site (U7.5; Fig. 2, lanes 5–7) and the one targeted to the 3′ splice site (U7.3; Fig. 2, lanes 8–10) corrected aberrant splicing of IVS2–705 pre-mRNA in a dose-dependent manner. Quantitative analysis of the results showed that at similar concentrations, the U7.3 and U7.5 RNAs corrected splicing to a similar level. At 2 μg/ml of DNA per 105 cells, the level of correct splicing was approximately 50% for both constructs. Note that visualization of the correct and aberrant PCR products overestimates the amount of aberrantly spliced RNA because it contains approximately twice as many labeled adenosine nucleotides (see Materials and Methods) as the correct one. As expected, transfection of the cells with the vector expressing anti-histone U7 snRNA (U7SmOPT) had no effect on splicing of IVS2–705 pre-mRNA (Fig. 2, lanes 2–4), confirming the sequence specificity of the observed antisense effects.
Figure 2.
Correction of aberrant splicing by U7 snRNA. Total RNA from HeLa IVS2–705 cell line transiently transfected with 0.5, 1, and 2 μg of U7 snRNA constructs (shown at the top) was analyzed by RT-PCR. Lanes: 1, untreated IVS2–705 cells; 2–4, cells transfected with U7SmOPT; 5–7 and 8–10, cells transfected with U7.5 and U7.3 constructs, respectively; 11, RNA from human blood. The sizes (in nucleotides) of PCR bands representing aberrantly and correctly spliced mRNAs are indicated on the left. The results of RT-PCR are also shown in Figs. 3, 4A, 5, 6A, and 7B. Quantification of these results took into account the approximately 2-fold higher [32P]dAp content of the aberrant PCR product than that of the correct one.
In an attempt to improve correction of splicing, we have introduced two additional modifications into the U7.3 construct. First, the antisense sequence was extended from 19 to 24 nt (U7.324, Fig. 1C), anticipating that the higher affinity of the longer sequence would increase the level of correct splicing. Second, because two of the nucleotides of the anti-globin sequence in U7.3 overlap with the Sm-binding site (Fig. 1C), it seemed possible that the bound Sm proteins might interfere with the antisense hybridization, reducing the correction of splicing. Hence, a 4-nt spacer was inserted between the SmOPT element and the antisense sequence in construct U7.34 (Fig. 1C).
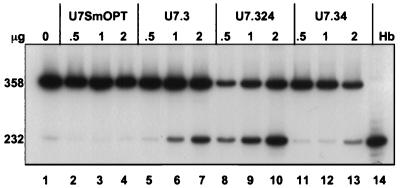
Transfection of the IVS2–705 cells with the U7.324 plasmid led to a significant increase of correct splicing (Fig. 3, lanes 8–10) relative to the unmodified U7.3 vector (Fig. 3, lanes 5–7). At 2 μg of vector DNA the level of correct splicing increased to 65% (Fig. 3, lane 10). This result suggests that extension of the antisense sequence improves the binding efficiency of the modified U7 snRNP. In contrast, addition of the 4-nt spacer in the U7.34 construct (Fig. 3, lanes 11–13) or a 10-nt spacer (data not shown) had no beneficial effect on correction of splicing and actually seemed to lead to a decrease in the correction of splicing when compared with the U7.3 vector. One interpretation of these results is that the Sm protein complex did not significantly interfere with the interactions between the 5′ end of the modified U7 snRNA and its target splice site. Additionally, the spacer sequence itself may have destabilized the U7 snRNA, decreasing its concentration, or modified its structure such that binding to its target splice site was less effective.
Figure 3.
Effect of modification of U7.3 RNA on correction of splicing. Lanes: 1, untreated IVS2–705 cells; 2–4, U7SmOPT; 5–7, U7.3; 8–10, U7.324; 11–13, U7.34. See Fig. 1C for sequences of the constructs. Lane 14, RNA from human blood. All other designations are as described in the legend to Fig. 2.
Immunoblotting with polyclonal antibody to human hemoglobin of protein from cells transiently transfected with U7.324 showed that the newly generated, correctly spliced β-globin mRNA was translated into full-length β-globin (Fig. 4). In agreement with RT-PCR results shown in Fig. 4A, cells with higher levels of correctly spliced β-globin mRNA contained increased amounts of full-length β-globin (Fig. 4B). Clearly, the generation of the β-globin protein was due to the effect of U7.324 snRNA on IVS2–705 pre-mRNA splicing.
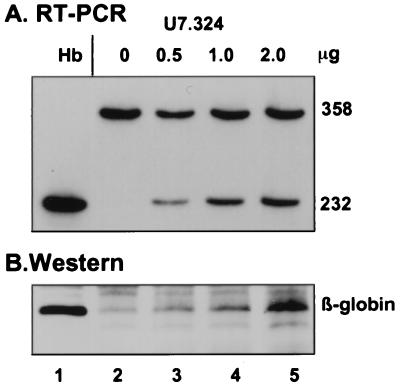
Figure 4.
Dose-dependent generation of β-globin mRNA (A) and protein (B) by U7.324. (A) Lanes: 1, RNA from human blood; 2–5, RNA from cells transfected with increasing amounts of U7.324 plasmid as indicated at the top. (B) Lanes: 1, protein from cell line expressing full-length β-globin protein; 2–5, total protein from cells transfected as in A. The proteins were analyzed by immunoblots probed with polyclonal anti-hemoglobin antibody (see Materials and Methods).
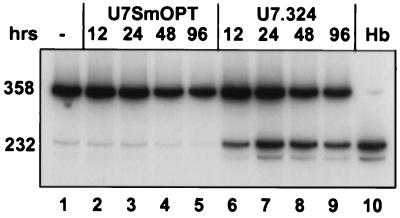
Fig. 5 shows the time course of the restoration of correct splicing of β-globin pre-mRNA after transient transfection of the IVS2–705 cell line with the U7.324 plasmid. RT-PCR analysis of the total RNA showed that a correction of splicing could be detected as early as 12 hr posttransfection (lane 6) and persisted through the 96-hr time point (lanes 7–9). Note that at 96 hr the transfected HeLa cells must have divided at least three to four times, and yet the level of splicing correction remained essentially unchanged. During the same time frame the treatment of cells with the U7SmOPT control construct had no effect on splicing of IVS2–705 pre-mRNA (lanes 2–5).
Figure 5.
Time course of correction of splicing. The RNA from IVS2–705 cells transfected with either U7SmOPT (lanes 2–5) or U7.324 (lanes 6–9) was isolated 12, 24, 48, and 96 hr posttransfection as indicated at the top of the figure. The control lanes (lanes 1 and 10) and all other designations are as described in the legend to Fig. 2.
Although in transient expression experiments the correction of splicing was evident for an extended period of time, the main advantage of the U7 vectors lies in their potential for permanent expression of antisense RNA and concomitant permanent correction of splicing. To test this possibility, stable cell lines were generated by cotransfecting IVS2–705 HeLa cells with the U7.324 vector and a plasmid carrying the hygromycin-resistance marker. Analysis of hygromycin-resistant colonies showed that several clones corrected IVS2–705 pre-mRNA splicing, albeit at different levels (Fig. 6A). In the most effective cell lines, the level of correction was 40–45% (Fig. 6A, lanes 3 and 4, respectively); Fig. 6A, lane 5, shows a poorly correcting cell line, with only approximately 15% of correctly spliced β-globin mRNA.
Additional experiments provided evidence that the correction of splicing in the selected cell lines is a consequence of the expression of U7.324 snRNA. The U7 RNA levels were measured directly by RT-PCR of total cellular RNA with U7-specific primers (Fig. 6B, lanes 6–9). Comparison of these results with those shown in Fig. 6A shows that the highest expression of U7.324 snRNA in cell line 705U7.324.4 (Fig. 6B, lane 8) correlates well with the highest level of correction observed in the same cell line (Fig. 6A, lane 4). The expression of U7.324 RNA in the remaining cell lines (Fig. 6B, lanes 7 and 9) also is commensurate with the correction of splicing (Fig. 6A, lanes 3 and 5, respectively). PCR analysis of the DNA from the selected cell lines shows that the differences in the level of U7.324 RNA expression are most likely due to different copy numbers of the U7 genes (Fig. 6B, lanes 3–5), because there is a correlation between the amounts of DNA amplification products and the levels of RNA expression and splicing correction. Finally, the possibility that the RT-PCR signal (Fig. 6B, lanes 7–9) may have originated from genomic DNA contamination of the isolated RNA was excluded by the absence of the U7-specific band (86 nt) when the reverse transcription step was omitted from the RT-PCR protocol (Fig. 6B, lanes 11–13). That PCR products were never detectable in the IVS2–705 parent cell line, which had not been transfected with the U7 vectors (Fig. 6B, lanes 2, 6, and 10), attests to the sequence specificity of the assays and eliminates the possibility that the 86-nt band was generated from endogenous human U7 genes.
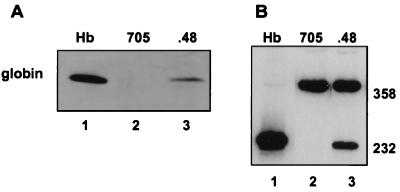
To ascertain that the stable transfection with U7 snRNA led not only to correction of splicing but also to stable expression of human β-globin, the protein lysates from another stable cell line, 705U7.324.48, were assayed by immunoblotting. The results showed significant accumulation of full-length β-globin protein (Fig. 7A, lane 3); accordingly, the RT-PCR analysis showed that the level of splicing correction in this cell line was approximately 55% (Fig. 7B, lane 3). The stably transfected cells appear to have growth rates comparable to that of the wild-type HeLa cells (data not shown), suggesting that expression of the modified U7 snRNA is not toxic to the cells.
Figure 7.
Correlation of β-globin protein (A) and mRNA (B) expression in stable cell line 705U7.324.48. (A) Western blot of protein from cell line expressing full-length β-globin protein (lane 1). Lanes: 2, protein from IVS2–705 cell line; 3, protein from cell line 705U7.324.48. (B) Lanes: 1, RNA from human blood; 2, RNA from IVS2–705 cell line; 3, RNA from cell line 705U7.324.48 stably expressing U7.324 snRNA.
To determine whether efficient correction of splicing relies on an excess of U7 snRNA over its target, the IVS2–705 pre-mRNA, their levels of expression in the 705U7.324.48 cell line were compared. RT-PCR analysis was performed on total cellular RNA by using the standard U7 primers and the β-globin primers that hybridize to exon 2 and intron 2 (see Fig. 1A and Materials and Methods). The latter primers detect solely β-globin pre-mRNA. The results were quantitated and normalized against standard curves obtained by PCR of IVS2–705 and U7.324 plasmid DNA. Interestingly, this analysis showed that there was an approximately 10-fold excess of β-globin pre-mRNA over U7 snRNA. This is not surprising, because transcription of the β-globin gene was driven by a strong cytomegalovirus promoter. We conclude that U7 snRNAs provide a specific and efficient mode of delivery of antisense sequences to the targeted splice sites.
DISCUSSION
The expression of U7 snRNA, modified to hybridize to aberrant splice sites in IVS2–705 thalassemic human β-globin pre-mRNA, reduced the incorrect splicing of pre-mRNA and led to increased levels of the correctly spliced mRNA and β-globin protein. U7 constructs antisense to either the novel 5′ splice site created by the 705 mutation (U7.5) or the cryptic 3′ splice site activated in the aberrant splicing pathway (U7.3 and its derivatives) were effective at restoring correct splicing. The cryptic 3′ splice site is utilized by the splicing machinery in IVS2–654, IVS2–705, and IVS2–745 thalassemic pre-mRNAs (28). Thus, the U7.324 construct should be useful for correction of splicing in all three mutants. Levels of correction reached 65% in transient expression and 55% in stable cell lines transfected with U7.324. Restoration of β-globin to these levels in thalassemic patients would be of therapeutic significance because transfusion therapy raises the hemoglobin to even lower levels yet improves the clinical status of the affected individuals (28).
The ability to generate cell lines in which the genetic defect that leads to incorrect splicing is by-passed and continuous production of a correct gene product is restored is highly encouraging. These results suggest a possibility of gene therapy based on the antisense concept. The patients’ bone marrow, in particular the erythroblasts and possibly the stem cells, could be transfected ex vivo with the antisense U7 vectors and reimplanted. Even if the expression of the U7 snRNA were short-lived, either because of lack of transfection of stem cells or promoter shut-off, both being common problems in the expression of transgenes (33, 34), the results may be relatively long lasting. This is because correction of β-globin pre-mRNA splicing driven by antisense U7 snRNA should increase the production of β-globin and reduce the imbalance between the α and β subunits of hemoglobin, consequently improving the survival of erythroblasts and promoting the maturation of erythrocytes. Because the lifespan of erythrocytes is approximately 120 days (35), the treated cells should persist in the bloodstream for an extended period of time.
The possibility of overexpression and/or inappropriate expression of the transfected gene constitutes serious concerns in gene therapy. In fact, overexpression of the β-globin transgene may lead to a new imbalance between α- and β-globin subunits and, conceivably, to symptoms of α-thalassemia. In this context, the correction of splicing by antisense U7 molecules offers an advantage because the β-globin subunits may at best reach the wild-type levels. Furthermore, even if the U7 snRNAs were inappropriately expressed in different cell types, their effects are expected to be limited only to cells that express the target sequence, β-globin pre-mRNA, i.e., to nucleated erythroblasts. The sequence specificity of the effect of U7 snRNAs targeted to the splice sites is substantiated by the negative results seen with the control U7SmOPT snRNA. It is further reinforced by the finding that the GenBank database of human sequences contains no sequence other than human β-globin intron 2 that corresponds to the 5′ and 3′ splice sites, even allowing for two mismatches.
For repair of a splicing mutation at the RNA level, it would be optimal to obtain high levels of expression of antisense RNA in the nucleus, where both expression of target pre-mRNAs and splicing occur. This requirement precludes the use of tRNAs as antisense vectors (36, 37), because this would result in accumulation of antisense sequences in the cytoplasm. In contrast, using U7 snRNA as an antisense carrier guarantees its nuclear localization, because the U7 snRNA will be transported from the cytoplasm to the nucleus in a manner similar to other Sm-type snRNAs. Because of their small size, secondary structure, and tight interactions with common Sm and other snRNP-specific proteins (38), the snRNAs, or rather their snRNP complexes, are very stable. In clinical applications the above properties would reduce the frequency of patient treatment. The modification of wild-type U7 snRNA to SmOPT, which was shown to increase its stability and nuclear uptake, in conjunction with its constitutive expression (30), clearly provided sufficient concentrations of the RNA to ensure efficient binding to the targeted splice sites and correction of splicing.
A possible concern about the practical application of our approach is that the level of expression of U7 snRNA may be insufficient to correct a significant fraction of β-globin mRNA, which is present in high levels in erythroid cells. However, the concentration of the actual U7 snRNA target, the β-globin pre-mRNA, is significantly lower. Furthermore, the finding that the 705U7.324.48 cell line expresses approximately 10-fold less U7 snRNA than β-globin pre-mRNA while maintaining a 55% level of correction indicates that a small amount of U7 snRNA is able to correct splicing in cells expressing large amounts of β-globin pre-mRNA. This is probably because the spliced-out β-globin intron 2, containing the U7 target, is rapidly degraded, whereas the stable U7 snRNP complex remains free to hybridize to another molecule of pre-mRNA. Boosting U7 snRNA expression through a multicopy plasmid or through use of viral vectors may be used to increase the levels of U7 snRNA in erythroid cells if they produce much greater quantities of β-globin pre-mRNA.
Other snRNAs have been used as convenient delivery agents for antisense therapeutics. Both U1 and U6 RNA have been modified as carriers of antisense sequences designed to down-regulate targeted sequences (39–43). U1 snRNA appears to be a particularly attractive candidate because it is known to bind to its target sequences, the 5′ splice sites, via a base-pairing mechanism. However, preliminary experiments showed that although a modified, transiently transfected U1 snRNA was efficiently transcribed, accounting for 25–30% of the total U1 RNA, its effect on splicing of the targeted adenovirus E1A or rabbit β-globin pre-mRNAs was minor (39). This may have been because of unstable binding of the 9-nt antisense sequence of the modified U1 RNA to its target, the inaccessibility of the target, or out-competition by wild-type U1 RNA. Interestingly, the anti-705 U7snRNA with its 24-nt antisense sequence was expressed at a level equal to that of endogenous U7 snRNA (U. Reber and D.S., data not shown). That and the concomitant lack of competition between the two molecules are likely to be responsible for the successful alteration of splicing reported here.
Because up to 15% of all point mutations in genetic diseases have been estimated to result in defective splicing (44), our approach may not be limited to thalassemic mutations. Furthermore, the same approach can be used to modify normal splicing patterns of constitutively and alternatively spliced pre-mRNAs resulting in changes in gene expression. Apart from the potential clinical applications, the ability to permanently modify splicing patterns of specific pre-mRNA may also prove useful in studies on the control of gene expression.
Acknowledgments
We thank Elizabeth Smith for technical assistance. This work was supported by National Institutes of Health Grant HL51940 to R.K. D.S. was supported by the Helmut Horten Stiftung, Roche Research Foundation, Hochschulstiftung Bern and by Grant 4037–44704 of the National Research Program 37 of the Swiss National Science Foundation.
ABBREVIATIONS
- IVS2
intron 2
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein
- RT-PCR
reverse transcription–PCR
References
- 1.Tolstoshev P. Annu Rev Pharm Toxicol. 1993;33:573–596. doi: 10.1146/annurev.pa.33.040193.003041. [DOI] [PubMed] [Google Scholar]
- 2.Fenjves E S, Schwartz P M, Blaese R M, Taichman L B. Hum Gene Ther. 1997;8:911–917. doi: 10.1089/hum.1997.8.8-911. [DOI] [PubMed] [Google Scholar]
- 3.Knowles M R, Hohneker K W, Zhou Z, Olsen J C, Noah T L, Hu P C, Leigh M W, Engelhardt J F, Edwards L J, Jones K R, et al. N Engl J Med. 1995;333:823–831. doi: 10.1056/NEJM199509283331302. [DOI] [PubMed] [Google Scholar]
- 4.Acsadi G, Dickson G, Love D R, Jani A, Walsh F S, Gurusinghe A, Wolff J A, Davies K E. Nature (London) 1991;352:815–818. doi: 10.1038/352815a0. [DOI] [PubMed] [Google Scholar]
- 5.Dunbar C, Kohn D. Hum Gene Ther. 1996;7:231–253. doi: 10.1089/hum.1996.7.2-231. [DOI] [PubMed] [Google Scholar]
- 6.Byun J, Kim S H, Kim J M, Yu S S, Robbins P D, Yim J, Kim S. Gene Ther. 1996;3:780–788. [PubMed] [Google Scholar]
- 7.Shi Q, Wang Y, Worton R. Hum Gene Ther. 1997;8:403–410. doi: 10.1089/hum.1997.8.4-403. [DOI] [PubMed] [Google Scholar]
- 8.Zhou S Z, Li Q, Stamatoyannopoulos G, Srivastava A. Gene Ther. 1996;3:223–229. [PubMed] [Google Scholar]
- 9.Ellis J, Pasceri P, Tan-Un K C, Wu X, Harper A, Fraser P, Grosveld F. Nucleic Acids Res. 1997;25:1296–1302. doi: 10.1093/nar/25.6.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadelain M, Wang C H, Antoniou M, Grosveld F, Mulligan R C. Proc Natl Acad Sci USA. 1995;92:6728–6732. doi: 10.1073/pnas.92.15.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole-Strauss A, Yoon K, Xiang Y, Byrne B C, Rice M C, Gryn J, Holloman W K, Kmiec E B. Science. 1996;273:1386–1389. doi: 10.1126/science.273.5280.1386. [DOI] [PubMed] [Google Scholar]
- 12.Perrine S P, Miller B A, Faller D V, Cohen R A, Vichinsky E P, Hurst D, Lubin B H, Papayannopoulou T. Blood. 1989;74:454–459. [PubMed] [Google Scholar]
- 13.Charache S, Barton F B, Moore R D, Terrin M L, Steinberg M H, Dover G J, Ballas S K, McMahon R P, Castro O, Orringer E P. Medicine. 1996;75:300–326. doi: 10.1097/00005792-199611000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Sher G D, Ginder G D, Little J, Yang S, Dover G J, Olivieri N F. N Engl J Med. 1995;332:1606–1610. doi: 10.1056/NEJM199506153322404. [DOI] [PubMed] [Google Scholar]
- 15.Collins A F, Pearson H A, Giardina P, McDonagh K T, Brusilow S W, Dover G J. Blood. 1995;85:43–49. [PubMed] [Google Scholar]
- 16.Sierakowska H, Sambade M J, Agrawal S, Kole R. Proc Natl Acad Sci USA. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominski Z, Kole R. Proc Natl Acad Sci USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith H O, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel M L. Proc Natl Acad Sci USA. 1991;88:9784–9788. doi: 10.1073/pnas.88.21.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galli G, Hofstetter H, Stunnenberg H G, Birnstiel M L. Cell. 1983;34:823–828. doi: 10.1016/0092-8674(83)90539-1. [DOI] [PubMed] [Google Scholar]
- 20.Birchmeier C, Schümperli D, Sconzo D, Birnstiel M L. Proc Natl Acad Sci USA. 1984;81:1057–1061. doi: 10.1073/pnas.81.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birnstiel M L, Schaufele F. In: Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Birnstiel M L, editor. Berlin: Springer; 1988. pp. 155–182. [Google Scholar]
- 22.Stefanovic B, Hackl W, Lührmann R, Schümperli D. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones D H, Howard B H. BioTechniques. 1991;10:62–66. [PubMed] [Google Scholar]
- 24.Jones D H, Winistorfer S C. BioTechniques. 1992;12:528–535. [PubMed] [Google Scholar]
- 25.Sierakowska H, Montague M, Agrawal S, Kole R. Nucleotides and Nucleosides. 1997;16:1173–1182. [Google Scholar]
- 26.Chen I T, Chasin L A. Mol Cell Biol. 1993;13:289–300. doi: 10.1128/mcb.13.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schagger H, von Jagov G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz E, Benz E J. In: Hematology: Basic Principles and Practice. Hoffman R, Benz E J, Shattil S J, Furie B, Cohen H J, editors. New York: Churchill Livingstone; 1991. pp. 368–392. [Google Scholar]
- 29.Bond U, Yario T A, Steitz J. Genes Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- 30.Spycher C, Streit A, Stefanovic B, Albrecht D, Konig T H, Schümperli D. Nucleic Acids Res. 1994;22:4023–4030. doi: 10.1093/nar/22.20.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimm C, Stefanovic B, Schümperli D. EMBO J. 1993;12:1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu C H, Murphy C, Gall J G. RNA. 1996;2:811–823. [PMC free article] [PubMed] [Google Scholar]
- 33.Page S M, Brownlee G G. J Invest Dermatol. 1997;109:139–145. doi: 10.1111/1523-1747.ep12319194. [DOI] [PubMed] [Google Scholar]
- 34.Palmer T D, Rosman G J, Osborne W R, Miller A D. Proc Natl Acad Sci USA. 1991;88:1330–1334. doi: 10.1073/pnas.88.4.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eadie G S, Brown I W, Curtis W G. J Clin Invest. 1955;34:629–637. doi: 10.1172/JCI103112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullenger B A, Lee T C, Smith C A, Ungers G E, Gilboa E. Mol Cell Biol. 1990;10:6512–6523. doi: 10.1128/mcb.10.12.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson J D, Ayers D F, Malmstrom T A, McKenzie T L, Ganousis L, Chowrira B M, Couture L, Stinchcomb D T. Nucleic Acids Res. 1995;23:2259–2268. doi: 10.1093/nar/23.12.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lührmann R, Kastner B, Bach M. Biochim Biophys Acta. 1990;1087:6255–6270. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 39.Yuo C, Weiner A M. Mol Cell Biol. 1989;9:3429–3437. doi: 10.1128/mcb.9.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montgomery R A, Dietz H C. Hum Mol Gen. 1997;6:519–525. doi: 10.1093/hmg/6.4.519. [DOI] [PubMed] [Google Scholar]
- 41.Good P D, Krikos A J, Li S X L, Bertrand E, Lee N S, Giver L, Ellington A, Zaia J A, Rossi J J, Engelke D R. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- 42.Liu D, Donegan J, Nuovo G, Mitra D, Laurence J. J Virol. 1997;71:4079–4085. doi: 10.1128/jvi.71.5.4079-4085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noonberg S B, Scott G K, Garavoy M R, Benz C C, Hunt C A. Nucleic Acids Res. 1994;22:2830–2836. doi: 10.1093/nar/22.14.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krawczak M, Reiss J, Cooper DN. Hum Gen. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]