Abstract
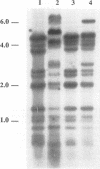
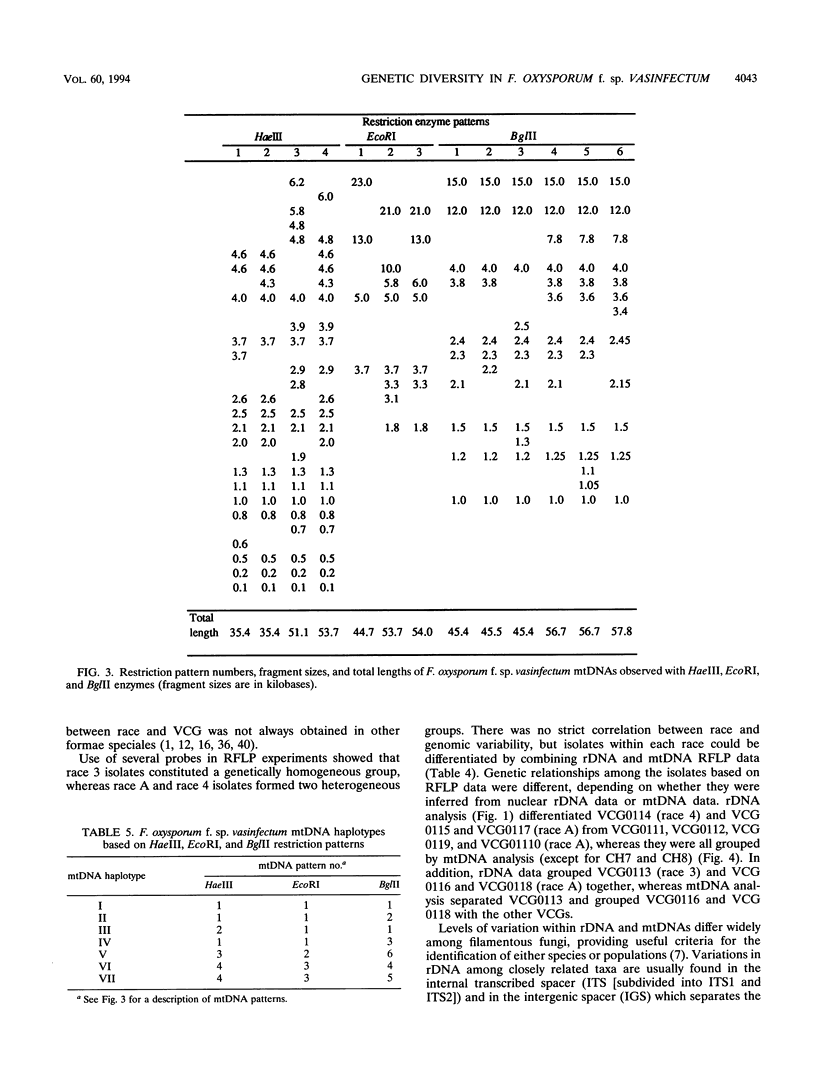
Restriction fragment length polymorphism (RFLP) and vegetative compatibility analyses were undertaken to assess genetic relationships among 52 isolates of Fusarium oxysporum f. sp. vasinfectum of worldwide origin and representing race A, 3, or 4 on cotton plants. Ten distinct vegetative compatibility groups (VCGs) were obtained, and isolates belonging to distinct races were never in the same VCG. Race A isolates were separated into eight VCGs, whereas isolates of race 3 were classified into a single VCG (0113), as were those of race 4 (0114). Ribosomal and mitochondrial DNA (rDNA and mtDNA) RFLPs separated four rDNA haplotypes and seven mtDNA haplotypes. Race A isolates displayed the most polymorphism, with three rDNA haplotypes and four mtDNA haplotypes; race 4 isolates formed a single rDNA group but exhibited three mtDNA haplotypes, while race 3 isolates had unique rDNA and mtDNA haplotypes. Two mtDNA molecules with distinct sizes were identified; the first (45-kb mtDNA) was found in all race A isolates and seven race 4 isolates, and the second (55-kb mtDNA) was found in all race 3 isolates and in two isolates of race 4. These two mtDNA molecules were closely related to mtDNAs of F. oxysporum isolates belonging to other formae speciales (conglutinans, lycopersici, matthioli, and raphani). Isolates within a VCG shared the same rDNA and mtDNA haplotypes, with the exception of VCG0114, in which three distinct mtDNA haplotypes were observed. Genetic relationships among isolates inferred from rDNA or mtDNA site restriction data were different, and there was not a strict correlation between race and RFLPs.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Kistler H. C., Benny U. The mitochondrial genome of Fusarium oxysporum. Plasmid. 1989 Jul;22(1):86–89. doi: 10.1016/0147-619x(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Marriott A. C., Archer S. A., Buck K. W. Mitochondrial DNA in Fusarium oxysporum is a 46.5 kilobase pair circular molecule. J Gen Microbiol. 1984 Nov;130(11):3001–3008. doi: 10.1099/00221287-130-11-3001. [DOI] [PubMed] [Google Scholar]
- Namiki F., Shiomi T., Kayamura T., Tsuge T. Characterization of the formae speciales of Fusarium oxysporum causing wilts of cucurbits by DNA fingerprinting with nuclear repetitive DNA sequences. Appl Environ Microbiol. 1994 Aug;60(8):2684–2691. doi: 10.1128/aem.60.8.2684-2691.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]