Abstract
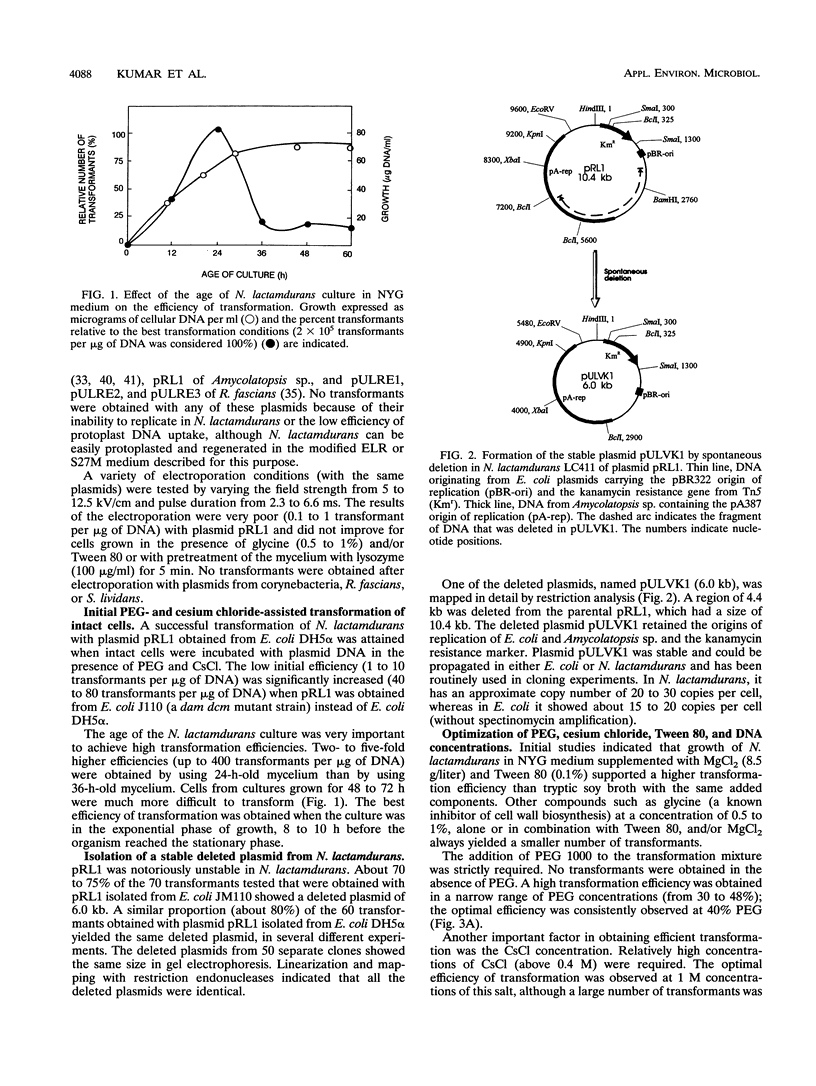
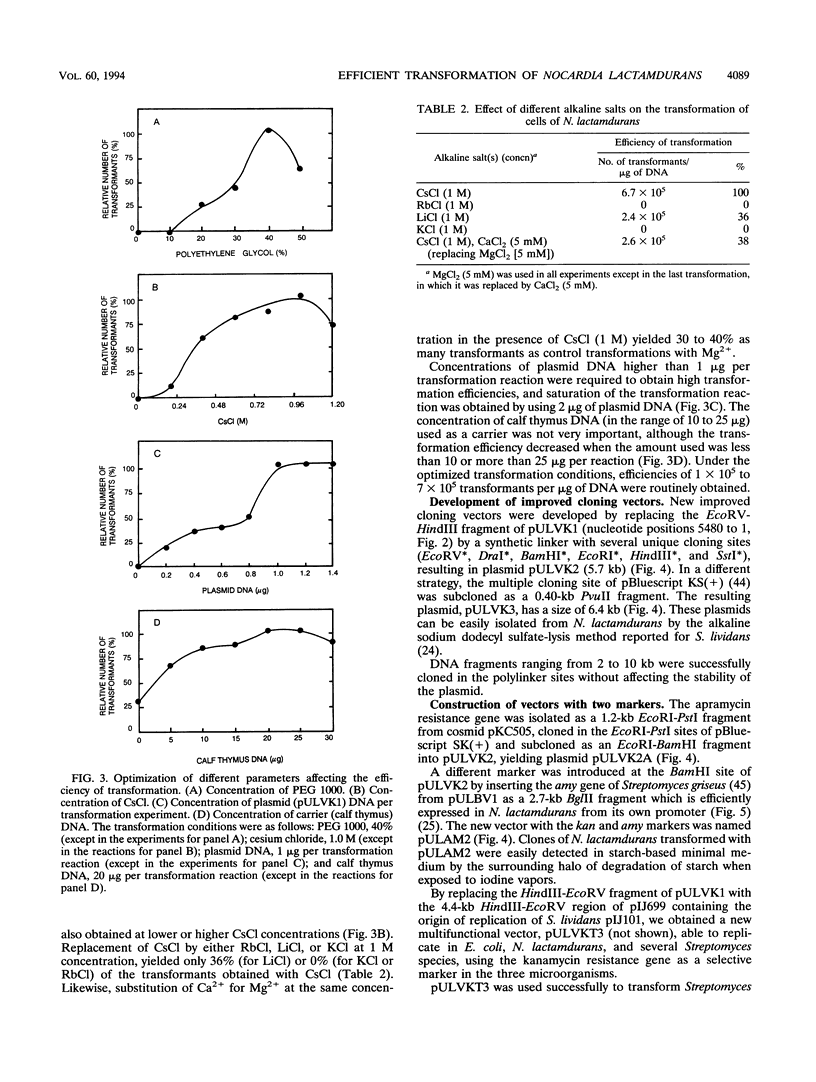
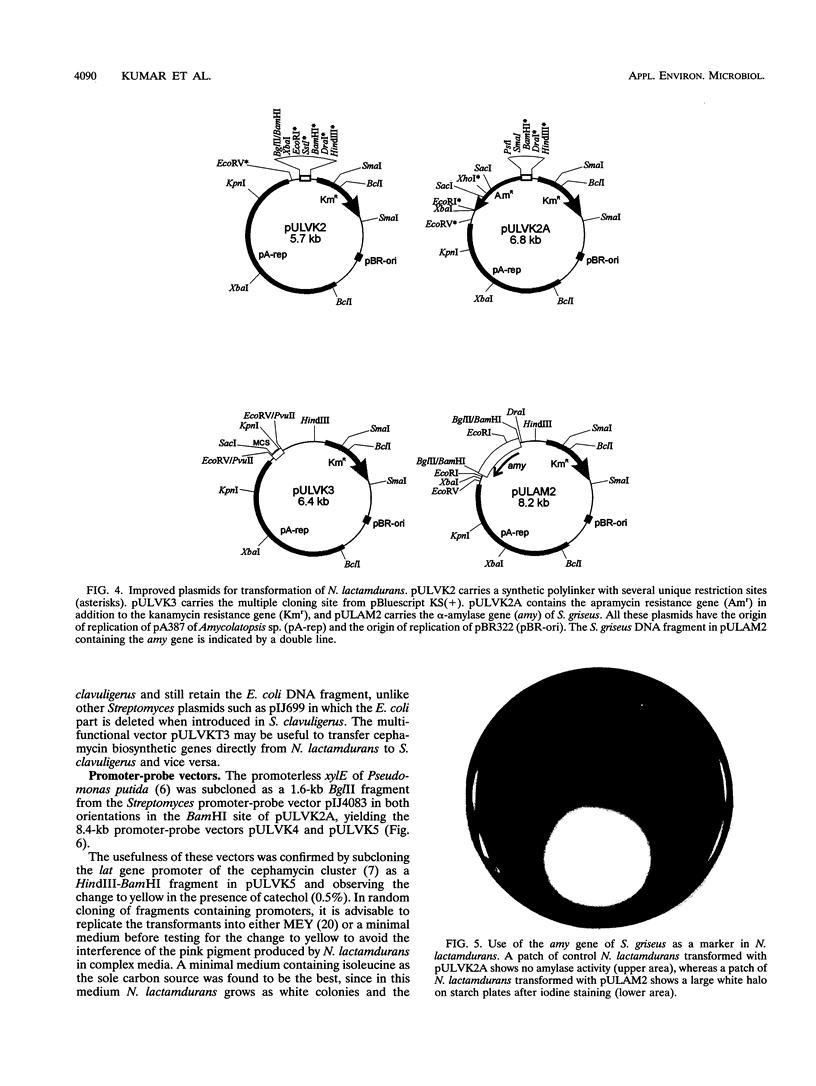
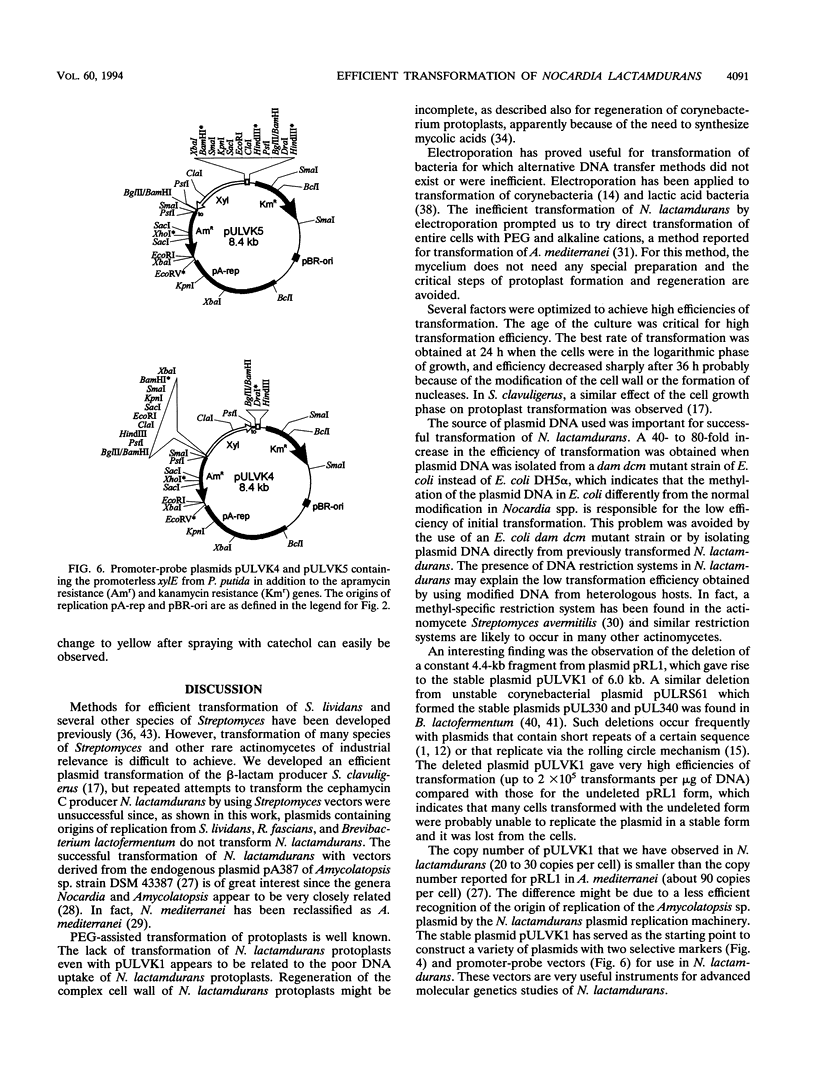
A high transformation efficiency (1 × 105 to 7 × 105 transformants per μg of DNA) of Nocardia lactamdurans LC411 was obtained by direct treatment of mycelium with polyethylene glycol 1000 and cesium chloride. A variety of vectors from Streptomyces lividans, Brevibacterium lactofermentum, Rhodococcus fascians, and a Nocardia (Amycolatopsis) sp. were tested; transformants could be obtained only with vectors derived from an endogenous plasmid of the Amycolatopsis sp. strain DSM 43387. Vectors carrying the kanamycin resistance gene (kan) as a selective marker were constructed. The transformation procedure has been optimized by using one of these vectors (pULVK1) and studying the influence of the age of the culture, concentrations of cesium chloride and polyethylene glycol, amount of plasmid DNA, and nutrient supplementations of the growth medium. Versatile shuttle cloning vectors (pULVK2 and pULVK3) have been developed by subcloning the pBluescript KS(+) multiple cloning site or a synthetic polylinker containing several unique restriction sites (EcoRV, DraI, BamHI, SstI, EcoRI, and HindIII). A second marker, the apramycin resistance gene (amr) has been added to the vectors (pULVK2A), allowing insertional inactivation of one of the markers while using the second one for selection. An alternative marker, the amy gene of Streptomyces griseus (pULAM2), which is easily detected by the release of extracellular amylase in transformants of N. lactamdurans carrying this vector, has been added. Two promoter-probe plasmids, pULVK4 and pULVK5, have been constructed, with the promoterless xylE gene as a reporter, for utilization in N. lactamdurans.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Castro J. M., Liras P., Laíz L., Cortés J., Martín J. F. Purification and characterization of the isopenicillin N synthase of Streptomyces lactamdurans. J Gen Microbiol. 1988 Jan;134(1):133–141. doi: 10.1099/00221287-134-1-133. [DOI] [PubMed] [Google Scholar]
- Clayton T. M., Bibb M. J. Streptomyces promoter-probe plasmids that utilise the xylE gene of Pseudomonas putida. Nucleic Acids Res. 1990 Feb 25;18(4):1077–1077. doi: 10.1093/nar/18.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque J. J., Liras P., Laiz L., Martín J. F. A gene encoding lysine 6-aminotransferase, which forms the beta-lactam precursor alpha-aminoadipic acid, is located in the cluster of cephamycin biosynthetic genes in Nocardia lactamdurans. J Bacteriol. 1991 Oct;173(19):6258–6264. doi: 10.1128/jb.173.19.6258-6264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque J. J., Liras P., Martín J. F. Genes for a beta-lactamase, a penicillin-binding protein and a transmembrane protein are clustered with the cephamycin biosynthetic genes in Nocardia lactamdurans. EMBO J. 1993 Feb;12(2):631–639. doi: 10.1002/j.1460-2075.1993.tb05696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coque J. J., Martín J. F., Calzada J. G., Liras P. The cephamycin biosynthetic genes pcbAB, encoding a large multidomain peptide synthetase, and pcbC of Nocardia lactamdurans are clustered together in an organization different from the same genes in Acremonium chrysogenum and Penicillium chrysogenum. Mol Microbiol. 1991 May;5(5):1125–1133. doi: 10.1111/j.1365-2958.1991.tb01885.x. [DOI] [PubMed] [Google Scholar]
- Coque J. J., Martín J. F., Liras P. Characterization and expression in Streptomyces lividans of cefD and cefE genes from Nocardia lactamdurans: the organization of the cephamycin gene cluster differs from that in Streptomyces clavuligerus. Mol Gen Genet. 1993 Jan;236(2-3):453–458. doi: 10.1007/BF00277148. [DOI] [PubMed] [Google Scholar]
- Cortés J., Martín J. F., Castro J. M., Láiz L., Liras P. Purification and characterization of a 2-oxoglutarate-linked ATP-independent deacetoxycephalosporin C synthase of Streptomyces lactamdurans. J Gen Microbiol. 1987 Nov;133(11):3165–3174. doi: 10.1099/00221287-133-11-3165. [DOI] [PubMed] [Google Scholar]
- Criado L. M., Martín J. F., Gil J. A. The pab gene of Streptomyces griseus, encoding p-aminobenzoic acid synthase, is located between genes possibly involved in candicidin biosynthesis. Gene. 1993 Apr 15;126(1):135–139. doi: 10.1016/0378-1119(93)90602-y. [DOI] [PubMed] [Google Scholar]
- Desomer J., Dhaese P., Van Montagu M. Conjugative transfer of cadmium resistance plasmids in Rhodococcus fascians strains. J Bacteriol. 1988 May;170(5):2401–2405. doi: 10.1128/jb.170.5.2401-2405.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez C., Cadenas R. F., Noirot-Gros M. F., Martin J. F., Gil J. A. Characterization of a region of plasmid pBL1 of Brevibacterium lactofermentum involved in replication via the rolling circle model. J Bacteriol. 1994 Jun;176(11):3154–3161. doi: 10.1128/jb.176.11.3154-3161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dominguez M., Martin J. F., Mahro B., Demain A. L., Liras P. Efficient plasmid transformation of the beta-lactam producer Streptomyces clavuligerus. Appl Environ Microbiol. 1987 Jun;53(6):1376–1381. doi: 10.1128/aem.53.6.1376-1381.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Domínguez M., Liras P., Martín J. F. Cloning and characterization of the isopenicillin N synthase gene of Streptomyces griseus NRRL 3851 and studies of expression and complementation of the cephamycin pathway in Streptomyces clavuligerus. Antimicrob Agents Chemother. 1991 Jan;35(1):44–52. doi: 10.1128/aac.35.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginther C. L. Sporulation and the production of serine protease and cephamycin C by Streptomyces lactamdurans. Antimicrob Agents Chemother. 1979 Apr;15(4):522–526. doi: 10.1128/aac.15.4.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen G. R., Bibb M. J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993 Feb 14;124(1):133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Kieser T., Melton R. E. Plasmid pIJ699, a multi-copy positive-selection vector for Streptomyces. Gene. 1988 May 15;65(1):83–91. doi: 10.1016/0378-1119(88)90419-2. [DOI] [PubMed] [Google Scholar]
- Lal R., Lal S., Grund E., Eichenlaub R. Construction of a hybrid plasmid capable of replication in Amycolatopsis mediterranei. Appl Environ Microbiol. 1991 Mar;57(3):665–671. doi: 10.1128/aem.57.3.665-671.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil D. J. Characterization of a unique methyl-specific restriction system in Streptomyces avermitilis. J Bacteriol. 1988 Dec;170(12):5607–5612. doi: 10.1128/jb.170.12.5607-5612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madoń J., Hütter R. Transformation system for Amycolatopsis (Nocardia) mediterranei: direct transformation of mycelium with plasmid DNA. J Bacteriol. 1991 Oct;173(20):6325–6331. doi: 10.1128/jb.173.20.6325-6331.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima P., Baltz R. H. Efficient plasmid transformation of Streptomyces ambofaciens and Streptomyces fradiae protoplasts. J Bacteriol. 1985 Jul;163(1):180–185. doi: 10.1128/jb.163.1.180-185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima P., McHenney M. A., Baltz R. H. Efficient transformation of Amycolatopsis orientalis (Nocardia orientalis) protoplasts by Streptomyces plasmids. J Bacteriol. 1987 May;169(5):2298–2300. doi: 10.1128/jb.169.5.2298-2300.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell Ian B., Achen Marc G., Hillier Alan J., Davidson Barrie E. A Simple and Rapid Method for Genetic Transformation of Lactic Streptococci by Electroporation. Appl Environ Microbiol. 1988 Mar;54(3):655–660. doi: 10.1128/aem.54.3.655-660.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. N., Richardson M. A., Kuhstoss S. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 1987;153:166–198. doi: 10.1016/0076-6879(87)53053-1. [DOI] [PubMed] [Google Scholar]
- Santamaria R. I., Gil J. A., Martin J. F. High-frequency transformation of Brevibacterium lactofermentum protoplasts by plasmid DNA. J Bacteriol. 1985 Apr;162(1):463–467. doi: 10.1128/jb.162.1.463-467.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría R. I., Martín J. F., Gil J. A. Identification of a promoter sequence in the plasmid pUL340 of Brevibacterium lactofermentum and construction of new cloning vectors for corynebacteria containing two selectable markers. Gene. 1987;56(2-3):199–208. doi: 10.1016/0378-1119(87)90137-5. [DOI] [PubMed] [Google Scholar]
- Stapley E. O., Jackson M., Hernandez S., Zimmerman S. B., Currie S. A., Mochales S., Mata J. M., Woodruff H. B., Hendlin D. Cephamycins, a new family of beta-lactam antibiotics. I. Production by actinomycetes, including Streptomyces lactamdurans sp. n. Antimicrob Agents Chemother. 1972 Sep;2(3):122–131. doi: 10.1128/aac.2.3.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Ward J. M., Hopwood D. A. Cloning of antibiotic resistance and nutritional genes in streptomycetes. J Bacteriol. 1982 Aug;151(2):668–677. doi: 10.1128/jb.151.2.668-677.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vigal T., Gil J. A., Daza A., García-González M. D., Martín J. F. Cloning, characterization and expression of an alpha-amylase gene from Streptomyces griseus IMRU3570. Mol Gen Genet. 1991 Feb;225(2):278–288. doi: 10.1007/BF00269860. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



