Abstract
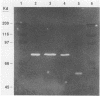
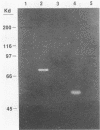
Streptococcus bovis is an important starch-degrading ruminal bacterium that has been implicated as being important in the etiology of a number of ruminal pathologies associated with diets high in grains. Previous studies with S. bovis have shown that amylase production was influenced by the growth substrate, but the nature of this regulation was not determined. The current study was conducted to better describe the regulatory phenomena and gain a better understanding of the molecular characteristics of this activity. Nutritional experiments demonstrated that the presence of starch or the starch-derived disaccharide maltose was required for maximum amylase production. Subsequent time-course experiments showed that amylase synthesis was induced by maltose and repressed by glucose, cellobiose, and fructose, while inulin and lactose had little effect on enzyme accumulation. The effects of the added antibiotics rifampin and tetracycline were consistent with transcriptional control of amylase synthesis. Analysis of S. bovis cells grown on glucose or maltose showed that they contained similar low levels of cyclic AMP, indicating that it was unlikely that regulation of amylase synthesis was mediated through a mechanism involving this nucleotide. The amylase gene from S. bovis JB1 was cloned and expressed in Escherichia coli. The amylase produced in E. coli was of lower molecular weight than that synthesized by S. bovis and had catalytic characteristics different from those of S. bovis amylase. When the gene was introduced back into S. bovis JB1, only one form of amylase activity was detected, indicating that the entire gene was present on this insert. The use of the amylase gene as a genetic probe for identification of S. bovis strains is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Band L., Henner D. J. Bacillus subtilis requires a "stringent" Shine-Dalgarno region for gene expression. DNA. 1984;3(1):17–21. doi: 10.1089/dna.1.1984.3.17. [DOI] [PubMed] [Google Scholar]
- Bounias M. N-(1-naphthyl)ethylenediamine dihydrochloride as a new reagent for nanomole quantification of sugars on thin-layer plates by a mathematical calibration process. Anal Biochem. 1980 Aug;106(2):291–295. doi: 10.1016/0003-2697(80)90523-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Clark R. G., Hu Y. J., Hynes M. F., Salmon R. K., Cheng K. J. Cloning and expression of an amylase gene from Streptococcus bovis in Escherichia coli. Arch Microbiol. 1992;157(3):201–204. doi: 10.1007/BF00245149. [DOI] [PubMed] [Google Scholar]
- Cotta M. A. Amylolytic activity of selected species of ruminal bacteria. Appl Environ Microbiol. 1988 Mar;54(3):772–776. doi: 10.1128/aem.54.3.772-776.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta M. A. Interaction of ruminal bacteria in the production and utilization of maltooligosaccharides from starch. Appl Environ Microbiol. 1992 Jan;58(1):48–54. doi: 10.1128/aem.58.1.48-54.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOBSON P. N., MACPHERSON M. Amylases of Clostridium butyricum and a Streptococcus isolated from the rumen of the sheep. Biochem J. 1952 Dec;52(4):671–679. doi: 10.1042/bj0520671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNGATE R. E., DOUGHERTY R. W., BRYANT M. P., CELLO R. M. Microbiological and physiological changes associated with acute indigestion in sheep. Cornell Vet. 1952 Oct;42(4):423–449. [PubMed] [Google Scholar]
- Hespell R. B., Wolf R., Bothast R. J. Fermentation of xylans by Butyrivibrio fibrisolvens and other ruminal bacteria. Appl Environ Microbiol. 1987 Dec;53(12):2849–2853. doi: 10.1128/aem.53.12.2849-2853.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylemon P. B., Phibbs P. V., Jr Evidence against the presence of cyclic AMP and related enzymes in selected strains of Bacteroides fragilis. Biochem Biophys Res Commun. 1974 Sep 9;60(1):88–95. doi: 10.1016/0006-291x(74)90176-4. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lacks S. A., Springhorn S. S. Renaturation of enzymes after polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate. J Biol Chem. 1980 Aug 10;255(15):7467–7473. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mackie R. I., Gilchrist F. M. Changes in Lactate-Producing and Lactate-Utilizing Bacteria in Relation to pH in the Rumen of Sheep During Stepwise Adaptation to a High-Concentrate Diet. Appl Environ Microbiol. 1979 Sep;38(3):422–430. doi: 10.1128/aem.38.3.422-430.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Tobian J. A., Jones K. R., Evans R. P., Clewell D. B. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene. 1982 Oct;19(3):345–353. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- Martin S. A., Russell J. B. Transport and phosphorylation of disaccharides by the ruminal bacterium Streptococcus bovis. Appl Environ Microbiol. 1987 Oct;53(10):2388–2393. doi: 10.1128/aem.53.10.2388-2393.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J. Regulation of sugar uptake and efflux in gram-positive bacteria. FEMS Microbiol Rev. 1989 Jun;5(1-2):149–156. doi: 10.1016/0168-6445(89)90019-3. [DOI] [PubMed] [Google Scholar]
- Russell J. B., Baldwin R. L. Comparison of maintenance energy expenditures and growth yields among several rumen bacteria grown on continuous culture. Appl Environ Microbiol. 1979 Mar;37(3):537–543. doi: 10.1128/aem.37.3.537-543.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Baldwin R. L. Comparison of substrate affinities among several rumen bacteria: a possible determinant of rumen bacterial competition. Appl Environ Microbiol. 1979 Mar;37(3):531–536. doi: 10.1128/aem.37.3.531-536.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Baldwin R. L. Substrate preferences in rumen bacteria: evidence of catabolite regulatory mechanisms. Appl Environ Microbiol. 1978 Aug;36(2):319–329. doi: 10.1128/aem.36.2.319-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Bottje W. G., Cotta M. A. Degradation of protein by mixed cultures of rumen bacteria: identification of Streptococcus bovis as an actively proteolytic rumen bacterium. J Anim Sci. 1981 Jul;53(1):242–252. doi: 10.2527/jas1981.531242x. [DOI] [PubMed] [Google Scholar]
- Russell J. B., Cotta M. A., Dombrowski D. B. Rumen Bacterial Competition in Continuous Culture: Streptococcus bovis Versus Megasphaera elsdenii. Appl Environ Microbiol. 1981 Jun;41(6):1394–1399. doi: 10.1128/aem.41.6.1394-1399.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B. Fermentation of cellodextrins by cellulolytic and noncellulolytic rumen bacteria. Appl Environ Microbiol. 1985 Mar;49(3):572–576. doi: 10.1128/aem.49.3.572-576.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B. Low-affinity, high-capacity system of glucose transport in the ruminal bacterium Streptococcus bovis: evidence for a mechanism of facilitated diffusion. Appl Environ Microbiol. 1990 Nov;56(11):3304–3307. doi: 10.1128/aem.56.11.3304-3307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Robinson P. H. Compositions and characteristics of strains of Streptococcus bovis. J Dairy Sci. 1984 Jul;67(7):1525–1531. doi: 10.3168/jds.S0022-0302(84)81471-X. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Saha B. C., Lecureux L. W., Zeikus J. G. Raw starch adsorption-desorption purification of a thermostable beta-amylase from Clostridium thermosulfurogenes. Anal Biochem. 1988 Dec;175(2):569–572. doi: 10.1016/0003-2697(88)90585-4. [DOI] [PubMed] [Google Scholar]
- Slyter L. L. Influence of acidosis on rumen function. J Anim Sci. 1976 Oct;43(4):910–929. doi: 10.2527/jas1976.434910x. [DOI] [PubMed] [Google Scholar]
- WALKER G. J. THE CELL-BOUND ALPHA-AMYLASES OF STREPTOCOCCUS BOVIS. Biochem J. 1965 Feb;94:289–298. doi: 10.1042/bj0940289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]