Abstract
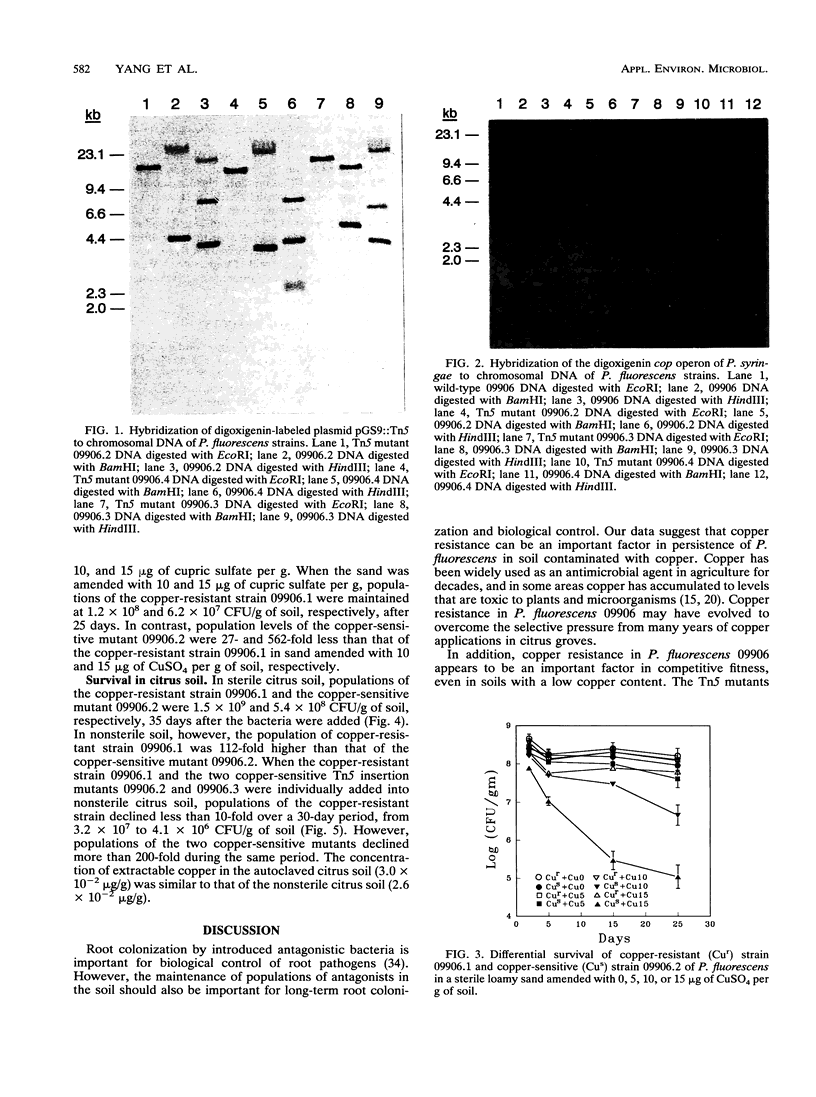
A copper-resistant strain (09906) of Pseudomonas fluorescens that was isolated from a citrus grove soil is being investigated as a biological control agent for Phytophthora root rot. Since citrus grove soils in California are often contaminated with copper from many years of copper fungicide applications, the role of copper resistance in survival of strain 09906 was investigated. Three copper-sensitive Tn5 mutants were obtained with insertions in different chromosomal DNA regions. These insertions were not in the chromosomal region that hybridized with the copper resistance operon (cop) cloned from Pseudomonas syringae. A copper-sensitive mutant survived as well as the wild type in a sterile loamy sand without added copper, but with 10 and 15 micrograms of CuSO4 added per g of soil, populations of the copper-sensitive mutant were 27- and 562-fold lower, respectively, than that of the wild type after a 25-day period. In a sterilized citrus grove soil, populations of the copper-sensitive mutant and wild-type strain were similar, but in nonsterile citrus soil, populations of the copper-sensitive mutant were 112-fold lower than the wild type after 35 days. These data suggest that copper resistance genes can be important factors in persistence of P. fluorescens in soil contaminated with copper. In addition, these genes appear to play a role in competitive fitness, even in soils with a low copper content.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender C. L., Cooksey D. A. Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1987 Feb;169(2):470–474. doi: 10.1128/jb.169.2.470-474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. L., Rouch D. A., Lee B. T. Copper resistance determinants in bacteria. Plasmid. 1992 Jan;27(1):41–51. doi: 10.1016/0147-619x(92)90005-u. [DOI] [PubMed] [Google Scholar]
- Casida L. E. Competitive ability and survival in soil of pseudomonas strain 679-2, a dominant, nonobligate bacterial predator of bacteria. Appl Environ Microbiol. 1992 Jan;58(1):32–37. doi: 10.1128/aem.58.1.32-37.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casida L. E. Relation to copper of N-1, a nonobligate bacterial predator. Appl Environ Microbiol. 1987 Jul;53(7):1515–1518. doi: 10.1128/aem.53.7.1515-1518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casida L. E. Response in Soil of Cupriavidus necator and Other Copper-Resistant Bacterial Predators of Bacteria to Addition of Water, Soluble Nutrients, Various Bacterial Species, or Bacillus thuringiensis Spores and Crystals. Appl Environ Microbiol. 1988 Sep;54(9):2161–2166. doi: 10.1128/aem.54.9.2161-2166.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J. S., Cooksey D. A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A., Azad H. R., Cha J. S., Lim C. K. Copper resistance gene homologs in pathogenic and saprophytic bacterial species from tomato. Appl Environ Microbiol. 1990 Feb;56(2):431–435. doi: 10.1128/aem.56.2.431-435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Induction of the copper resistance operon from Pseudomonas syringae. J Bacteriol. 1988 Sep;170(9):4399–4401. doi: 10.1128/jb.170.9.4399-4401.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1988 Jun;170(6):2879–2883. doi: 10.1128/jb.170.6.2879-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. D., Bhave M. R., Mercer J. F., Camakaris J., Lee B. T. Cloning and characterization of cutE, a gene involved in copper transport in Escherichia coli. J Bacteriol. 1991 Nov;173(21):6742–6748. doi: 10.1128/jb.173.21.6742-6748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouch D., Camakaris J., Lee B. T., Luke R. K. Inducible plasmid-mediated copper resistance in Escherichia coli. J Gen Microbiol. 1985 Apr;131(4):939–943. doi: 10.1099/00221287-131-4-939. [DOI] [PubMed] [Google Scholar]
- Selvaraj G., Iyer V. N. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol. 1983 Dec;156(3):1292–1300. doi: 10.1128/jb.156.3.1292-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992 Mar;56(1):195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevors J. T. Copper resistance in bacteria. Microbiol Sci. 1987 Jan;4(1):29–31. [PubMed] [Google Scholar]