Abstract
Cdc25C-associated kinase 1 (C-TAK1) has been implicated in cell cycle regulation and Ras signaling through its interactions with two putative substrates, the Cdc25C phosphatase and the MAPK scaffold KSR1. Here, we identify sequence motifs required for stable C-TAK1 association and substrate phosphorylation. Using a mutational approach to disrupt binding of C-TAK1 to KSR1 and Cdc25C, we demonstrate that C-TAK1 contributes to the regulation of these proteins in vivo through the generation of 14-3-3-binding sites. KSR1 proteins defective in C-TAK1 binding had severely reduced phosphorylation at the 14-3-3-binding site in vivo, were constitutively localized to the plasma membrane and had increased biological activity. Disruption of the Cdc25C–C-TAK1 interaction resulted in reduced 14-3-3-binding site phosphorylation and nuclear accumulation of Cdc25C in interphase cells. Finally, utilizing the acquired C-TAK1 binding and substrate phosphorylation data, we identify plakophilin 2 (PKP2) as a novel C-TAK1 substrate. Phosphorylation of PKP2 by C-TAK1 also generates a 14-3-3-binding site that influences PKP2 localization. These findings underscore the importance of C-TAK1 as a regulator of 14-3-3 binding and protein localization.
Keywords: 14-3-3/C-TAK1/phosphorylation motif/protein kinase/subcellular protein localization
Introduction
Intracellular protein trafficking plays an important role in the regulation of many cellular processes, including signal transduction, cell cycle regulation and apoptosis. In these processes, key components are often maintained as inactive factors in a different subcellular compartment from where they exert their effects. Only in response to the appropriate cellular cues do these molecules localize to their site of action. Two important mechanisms by which protein localization can be modulated are through protein interactions and phosphorylation events. A striking example of how protein interactions can affect localization is demonstrated by the Ras GTPase. In response to many extracellular stimuli, conversion of membrane-bound Ras to its GTP-bound state mediates the translocation of key cytoplasmic effectors, such as the Raf-1 kinase, to the cell surface (Marshall, 1996). Likewise, phosphorylation events can influence the movement of proteins from one cellular compartment to another. This mode of regulation is well illustrated by the binding of cytoplasmic SH2- and PTB-containing molecules to phosphotyrosine residues found on receptor tyrosine kinases at the cell surface (Yaffe, 2002b).
More recently, it has become apparent that phosphorylation-dependent binding of 14-3-3 molecules also plays a key role in regulating intracellular protein movement. 14-3-3 family members are highly conserved dimeric proteins that bind mainly to phosphoserine motifs (Muslin et al., 1996; Yaffe et al., 1997). The primary manner by which 14-3-3 affects protein localization is through molecular interference, in that the binding of 14-3-3 directly alters the functioning of nearby subcellular targeting sequences found on the binding partner (Muslin and Xing, 2000; Yaffe, 2002a). 14-3-3 has been shown to modulate the nuclear/cytoplasmic shuttling of numerous proteins, including forkhead (Brunet et al., 1999), Cdc25C (Kumagai and Dunphy, 1999; Yang et al., 1999; Graves et al., 2001) and histone deacetylase (Grozinger and Schreiber, 2000; Kao et al., 2001). Similarly, 14-3-3 has been found to play a regulatory role in the cytoplasmic/membrane shuttling of proteins, such as the Raf-1 kinase (Jaumot and Hancock, 2001; Kubicek et al., 2002; Light et al., 2002) and the mitogen-activated protein kinase (MAPK) scaffolding protein KSR1 (Müller et al., 2001). With respect to KSR1, the binding of 14-3-3 molecules to two phosphoserine residues (S297 and S392) is critical for maintaining the cytoplasmic localization of the KSR1 complex in quiescent cells (Müller et al., 2001). In response to stimuli, the phosphorylation state of S392 is reduced, releasing 14-3-3 from this site. The removal of 14-3-3 appears to expose the cysteine-rich C1 domain of KSR1, which then directs the translocation of KSR1 to the plasma membrane, where it facilitates signal transmission from Raf-1 to MEK and MAPK (Müller et al., 2001; Zhou et al., 2002).
In searching for kinases that might regulate the phosphorylation state of KSR1, we recently found that the protein kinase Cdc25C-associated kinase 1 (C-TAK1) is a constitutive component of the KSR1 complex (Müller et al., 2001). KSR1 itself is a substrate of C-TAK1 in vitro, and the KSR1 residue phosphorylated by C-TAK1 is the S392/14-3-3-binding site. C-TAK1 is a member of the EMK/MARK/Par1 kinase family and was first cloned based on its ability to associate with and phosphorylate Cdc25C (Ogg et al., 1994; Peng et al., 1998). Subsequently, the tyrosine phosphatase PTPH1 was found to be phosphorylated by C-TAK1 in vitro (Zhang et al., 1997). Strikingly, for all known substrates of C-TAK1 (KSR1, Cdc25C and PTPH1), the residue phosphorylated by C-TAK1 serves as a 14-3-3-binding site.
To determine if the regulation of 14-3-3 binding is a general feature of C-TAK1 function and to uncover other proteins that might be substrates of C-TAK1, we initiated a mutagenesis study to define sequence motifs required for C-TAK1 binding and substrate phosphorylation. Here we report that a hydrophobic residue found at the +5 position relative to the site phosphorylated by C-TAK1 is critical for the stable association of C-TAK1 with its previously defined substrates Cdc25C and KSR1. Moreover, we find that hydrophobic residues at the –5, +1 and +5 positions as well as an arginine at the –3 position are required for optimal C-TAK1 phosphorylation. Analysis of KSR1 and Cdc25C proteins defective in C-TAK1 binding revealed the involvement of C-TAK1 in regulating the phosphorylation state and subcellular localization of these proteins, findings confirmed by overexpression of a dominant-inhibitory C-TAK1 protein. Finally, by searching protein databases with the acquired C-TAK1 binding and substrate phosphorylation data, we identify plakophilin 2 (PKP2) as a novel C-TAK1 substrate and show that C-TAK1-mediated phosphorylation of PKP2 generates a 14-3-3-binding site, which in turn influences the intracellular localization of PKP2.
Results
Defining KSR1 sequence(s) required for C-TAK1 binding
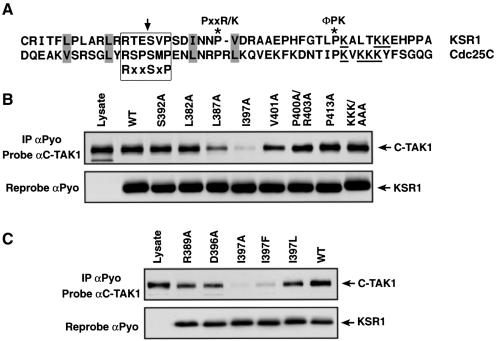
C-TAK1 has been shown to stably associate with two of its substrate proteins, KSR1 and Cdc25C. Experiments analyzing the binding of C-TAK1 to various KSR1 and Cdc25C deletion mutants have revealed that the interaction with C-TAK1 requires residues 377–424 of murine KSR1 (Müller et al., 2001; data not shown) and residues 200–256 of human Cdc25C (Peng et al., 1998). When these regions of KSR1 and Cdc25C are compared, several areas of amino acid similarity can be observed (Figure 1A). In addition to the C-TAK1 target phosphorylation site that confers 14-3-3 binding (S392 for KSR1 and S216 of Cdc25C), these regions contain a cluster of lysine residues, proline residues found in specific sequence contexts [PxxR/K; a hydrophobic residue (Φ)PK] and hydrophobic residues located at similar positions surrounding the target phosphorylation site. To determine whether any of these amino acids are required for the interaction with C-TAK1, we first generated KSR1 proteins that have mutations in these sites. For this analysis, the mutations were incorporated into a KSR1 protein encoding the N-terminal 424 amino acids of KSR1 (KSR1/N′424), as this protein contains the binding site for C-TAK1, but not the interaction sites for many of the known KSR1-associated proteins, such as Raf-1, MEK, ERK, Hsp90 and Cdc37 (Morrison, 2001). The KSR1 mutants were then transiently expressed in Cos cells and examined for their ability to interact with endogenous C-TAK1. As shown in Figure 1B, C-TAK1 strongly associated with both the wild-type N′424 protein and the S392A mutant, indicating that the target site of phosphorylation is not required for C-TAK1 binding. In addition, the KSR1–C-TAK1 interaction was not altered by mutation of the lysine cluster, the proline motifs or the L383 and V401 hydrophobic residues. Strikingly, while C-TAK1 binding was somewhat reduced for the L387A mutant, little to no C-TAK1 was associated with the I397A mutant, indicating the importance of this residue for the stable interaction between KSR1 and C-TAK1.
Fig. 1. KSR1 residue(s) required for C-TAK1 binding. (A) Amino acids 201–249 of Cdc25C and 377–424 of KSR1. Regions of amino acid similarity are depicted in the following manner: the cluster of lysine residues is underlined, conserved hydrophobic residues are highlighted, and proline residues found in specific sequence contexts (PxxR and ΦPK) are indicated with asterisks. The serine residue phosphorylated by C-TAK1 is denoted with an arrow, and the 14-3-3-binding motif (RxxSxP) is boxed. (B and C) Wild-type and KSR1/N′424 mutant proteins were immunoprecipitated from Cos cell lysates, and the immune complexes examined by immunoblot analysis using C-TAK1 and Pyo antibodies.
Defining KSR1 sequences required for C-TAK1-mediated phosphorylation
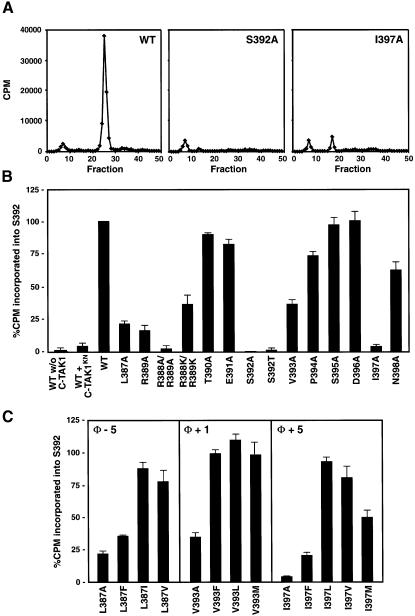
Due to the close proximity of the I397 residue to the site phosphorylated by C-TAK1, we next performed experiments to determine whether this or other residues surrounding the S392 phosphoacceptor site are required for recognition of KSR1 as a C-TAK1 substrate. For these experiments, a series of KSR1 N′424 proteins was generated in which each residue from amino acid 387 to 398 was mutated to alanine. The mutant proteins were expressed in Cos cells and affinity purified from cell lysates under conditions known to disrupt KSR1 protein interactions (Müller et al., 2001). The purified mutants were then incubated in vitro with purified C-TAK1 in the presence of [γ-32P]ATP. The labeled proteins were isolated, digested with trypsin, and analyzed for S392 phosphorylation by high-pressure liquid chromatography (HPLC) (Figure 2A and B). In these assays, phosphorylation of wild-type N′424 was only observed in the presence of kinase-active C-TAK1, indicating that the purified KSR1 and C-TAK1 proteins did not contain contaminating kinase activities. Wild-type N′424 was phosphorylated efficiently by C-TAK1 and, as expected, phosphorylation was abolished by mutation of the S392 site. In addition, mutation of R389 severely reduced S392 phosphorylation, consistent with the fact that C-TAK1 is a member of the CAMK group of kinases, whose consensus phosphorylation motif often includes a basic residue at the –3 position relative to the phosphoacceptor site (Pearson and Kemp, 1991; Songyang et al., 1996). Likewise, mutation of hydrophobic residues at the –5 (L387A), +1(V393A) and +5 (I397A) positions inhibited C-TAK1-mediated S392 phosphorylation, with mutation of the +5 site having the most severe effect. Further mutational analysis revealed that while any hydrophobic residue was tolerated at the +1 position, hydrophobic residues with aliphatic side chains (I, L and V) were preferred at the –5 and +5 positions (Figure 2C). When the L387A–N398A panel of KSR1/N′424 mutants were assayed for their ability to stably associate with C-TAK1, only mutations that altered the I397 site were found to significantly disrupt C-TAK1 binding (Figure 1C, and data not shown). Based on this analysis, we conclude that C-TAK1 is a basophilic kinase, and that hydrophobic residues surrounding the phosphoacceptor site, in particular at positions –5, +1 and +5, play a critical role in C-TAK1 substrate phosphorylation. Interestingly, other members of the CAMK group, such as SNF1, AMP-activated kinase (AMPK), calcium/calmodulin-dependent kinase I (CAMKI), Chk1 and Chk2, have also been found to have a strong preference for substrates containing hydrophobic residues at the –5 position and a weaker requirement for hydrophobic residues at the +1 position (Dale et al., 1995; Hutchins et al., 2000; O’Neill et al., 2002). However, C-TAK1 appears to be unique among CAMK members in its preference for a hydrophobic amino acid at the +5 position, which, significantly, is also the residue required for the stable interaction between KSR1 and C-TAK1.
Fig. 2. Identification of KSR1 residues required for C-TAK1 phosphorylation. (A) In vitro kinase assays were performed using purified wild-type C-TAK1 or kinase-negative C-TAK1 (C-TAK1KN) and 0.2 µg of the purified KSR1/N′424 proteins. The labeled KSR1 proteins were resolved by SDS–PAGE, isolated from the gel matrix and digested with trypsin. The tryptic phosphopeptides were then separated by reverse-phase HPLC. The profile of radioactivity collected in the HPLC column fractions is shown for wild-type, S392A and I397A KSR1/N′424 proteins. (B and C) Kinase assays and HPLC analysis were performed as in (A) and the radioactivity incorporated into S392 was quantitated. S392 phosphorylation of wild-type KSR1/N′424 was considered to be 100% and the level of S392 phosphorylation for each mutant was compared with wild-type.
Residues of Cdc25C required for the C-TAK1 interaction
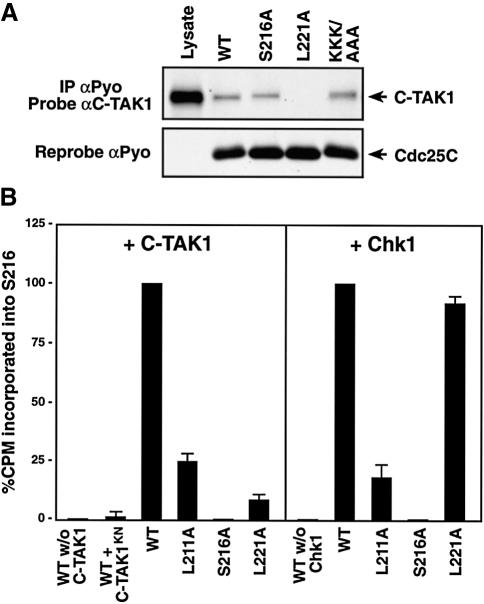
To determine whether a hydrophobic residue at the +5 position relative to the C-TAK1 phosphoacceptor site is required for stable binding of C-TAK1 to other substrates, we examined what effect mutation of the analogous Cdc25C residue (L221) would have on the Cdc25C–C-TAK1 interaction (Figure 3A). Polyomavirus (Pyo)-tagged Cdc25C proteins were generated in which the S216 phosphorylation site (S216A), the lysine residues at positions 241, 242 and 243 (KKK/AAA), and the hydrophobic residue at position 221 (L221A) were mutated to alanine. When the ability of these proteins to associate with endogenous C-TAK1 was examined, we found that equivalent amounts of C-TAK1 were observed in the wild-type, S216A and KKK/AAA Cdc25C immune complexes; however, no C-TAK1 was detected in immunoprecipitates of L221A Cdc25C. Thus, as with KSR1, the hydrophobic residue at the +5 position relative to the C-TAK1 phosphoacceptor site plays a critical role in mediating the interaction between C-TAK1 and Cdc25C.
Fig. 3. Analysis of Cdc25C residues required for C-TAK1-mediated phosphorylation. (A) Cos cells were transfected with constructs encoding Pyo-tagged wild-type, S216A, L221A or K242A/K243A/K244A (KKK/AAA) Cdc25C. Proteins were immunoprecipitated from cell lysates, and the immune complexes were examined by immunoblot analysis using C-TAK1 and Pyo antibodies. (B) In vitro phosphorylation of purified Cdc25C proteins by purified C-TAK1 and Chk1. For each mutant, the phosphate incorporated into the S216 site was quantitated and compared with wild-type.
To investigate whether the L221 residue is also required for the phosphorylation of Cdc25C by C-TAK1, in vitro kinase assays were performed using purified Cdc25C and C-TAK1 proteins (Figure 3B). For this analysis, wild-type Cdc25C and proteins containing a mutation in either S216, L221 or the hydrophobic residue at the –5 position (L211A) were examined. In addition, because the Chk1 kinase has also been found to phosphorylate Cdc25C on the S216 site (Peng et al., 1997; Sanchez et al., 1997), the Cdc25C proteins were assayed for their ability to be phosphorylated by purified Chk1. HPLC analysis of the labeled proteins revealed that both kinases were able to phosphorylate wild-type Cdc25C on S216. The L211A mutant was a poor substrate for either C-TAK1 or Chk1, indicating the importance of a hydrophobic residue at the –5 position for substrate phosphorylation by both kinases. In contrast, mutation of L221 only disrupted C-TAK1-mediated phosphorylation of S216. These findings are consistent with the unique requirement of C-TAK1 for a hydrophobic residue at the +5 position, and further demonstrate the importance of this residue for both C-TAK1 binding and substrate phosphorylation.
Functional assessment of the KSR1–C-TAK1 interaction in vivo
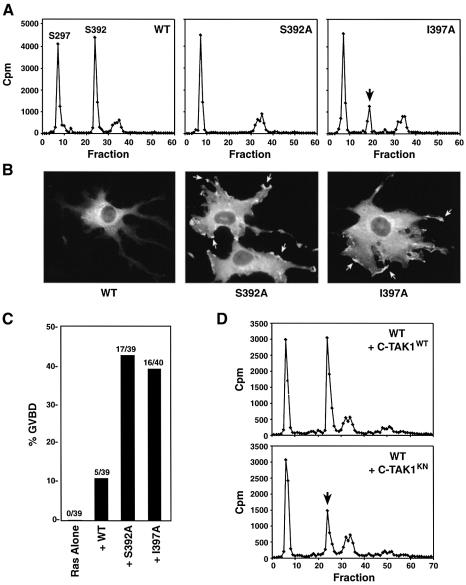
The identification of mutations that disrupt the interaction of C-TAK1 with its substrates allows us to address the functional importance of C-TAK1 binding in vivo. As shown above, mutation of the hydrophobic residue at the +5 position relative to the phosphoacceptor site severely disrupts the binding and in vitro phosphorylation of KSR1 and Cdc25C by C-TAK1; however, based upon known phosphorylation recognition motifs and data presented in Figure 3B, this mutation should not interfere with the potential ability of other basophilic kinases, such as members of the AGC and CAMK families, to phosphorylate these proteins. Therefore, to address the importance of C-TAK1 in mediating the phosphorylation of KSR1 on S392 in intact cells, we examined the in vivo phosphorylation state, subcellular localization and biological activity of a full-length KSR1 protein containing the I397A mutation. Previously, we have shown that KSR1 is constitutively phosphorylated on S392 in quiescent cells, and that mutation of the S392 site leads to loss of 14-3-3 binding, constitutive plasma membrane localization of KSR1 in cultured cells and enhanced Ras-dependent maturation of Xenopus oocytes (Cacace et al., 1999; Müller et al., 2001). As shown in Figure 4A, when the in vivo phosphorylation state of wild-type, S392A and I397A KSR1 proteins was examined by HPLC analysis, wild-type KSR1 was found to be phosphorylated on two major sites in quiescent cells, S297 (eluting in fraction 7) and S392 (eluting in fraction 24). Although mutation of I397 had no effect on S297 phosphorylation, phosphorylation of S392 was reduced ∼75%. Moreover, we found that this altered phosphorylation status was accompanied by constitutive localization of the I397A mutant to the plasma membrane in NIH-3T3 cells (Figure 4B), as well as accelerated Ras-dependent maturation kinetics in the Xenopus oocyte assay (Figure 4C). To address the role of C-TAK1 in S392 phosphorylation further, we next examined the in vivo phosphorylation state of wild-type KSR1 in cells overexpressing either wild-type C-TAK1 or a kinase-negative C-TAK1 protein, which has been shown to interfere with the activity of endogenous C-TAK1 (Sun et al., 2001). As depicted in Figure 4D, expression of the kinase-negative C-TAK1 specifically reduced phosphorylation of the S392 site. Taken together, these findings demonstrate that C-TAK1 is a primary in vivo kinase that phosphorylates KSR1 on S392.
Fig. 4. In vivo analysis of the KSR1–C-TAK1 interaction. (A) Quiescent Cos cells expressing wild-type, S392A or I397A KSR1 were labeled in vivo with [32P]orthophosphate. The labeled KSR1 proteins were isolated, digested with trypsin and examined by HPLC analysis. The arrow indicates the position of the peak containing the S392 peptide. It should be noted that the I397A substitution causes the S392 peptide to elute earlier in the HPLC gradient (as confirmed by sequence analysis). (B) NIH-3T3 cells were transfected with constructs encoding either wild-type, S392A or I397A KSR1. Localization of the KSR1 proteins was then determined by indirect immunofluorescence using the Pyo antibody. (C) Stage VI Xenopus oocytes were injected with RNAs encoding wild-type, S392A or I397A KSR1. Oocytes subsequently were injected with activated RasV12 RNA and scored for germinal vesicle breakdown (GVBD) 5 h following Ras injection. (D) Quiescent Cos cells co-expressing wild-type KSR1 and either wild-type C-TAK1 or kinase-negative C-TAK1 (C-TAK1KN) were labeled in vivo with [32P]orthophosphate and analyzed as described in (A).
Disruption of C-TAK1 binding alters the in vivo phosphorylation state and subcellular localization of Cdc25C
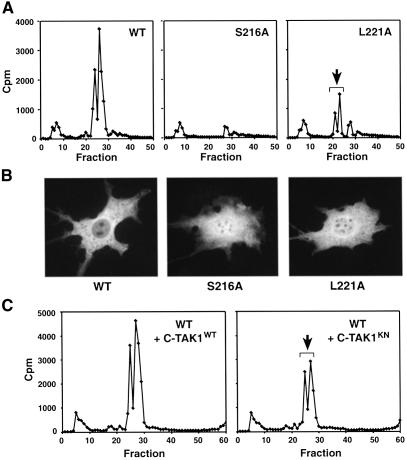
To examine whether C-TAK1 contributes to the phosphorylation of Cdc25C on S216 in vivo, we examined the phosphorylation state of wild-type, S216A or L221A Cdc25C proteins in quiescent interphase cells. HPLC analysis of the labeled proteins revealed that wild-type Cdc25C contains tryptic phosphopeptides that elute in fractions 24 and 26. Due to incomplete trypsin digestion at closely spaced lysine residues, K225 and K227, two S216-containing peptides are generated that elute in fractions 24 and 26. By sequence analysis, both fractions contain peptides phosphorylated at S216 and are not detected if the S216 site is mutated (Figure 5A). When the HPLC profile of L221A Cdc25C was examined, phosphorylation of S216 was found to be reduced ∼65%. In addition, overexpression of kinase-inactive C-TAK1 was also found to reduce the in vivo phosphorylation state of S216 (Figure 5C).
Fig. 5. In vivo analysis of the Cdc25C–C-TAK1 interaction. (A) Quiescent Cos cells expressing wild-type, S216A or L221A Cdc25C were labeled in vivo with [32P]orthophosphate. The labeled Cdc25C proteins were isolated, digested with trypsin and examined by HPLC analysis. (B) NIH-3T3 cells were transfected with constructs encoding Pyo-tagged wild-type, S216A or L221A Cdc25C. Localization of the Cdc25C proteins was then determined by indirect immunofluorescence using the Pyo antibody. (C) Quiescent Cos cells co-expressing wild-type Cdc25C and either wild-type C-TAK1 or kinase-negative C-TAK1 (C-TAK1KN) were labeled in vivo with [32P]orthophosphate and analyzed as described in (A).
Because S216 phosphorylation and associated 14-3-3 binding has been shown to regulate the nuclear–cytoplasmic shuttling of Cdc25C (Peng et al., 1998; Kumagai and Dunphy, 1999; Yang et al., 1999; Graves et al., 2001), we next examined what effect disruption of the Cdc25C– C-TAK1 interaction would have on the subcellular localization of Cdc25C (Figure 5B). As expected, wild-type Cdc25C, which is phosphorylated on S216, was localized exclusively in the cytosol. In contrast, both the S216A and L221A mutants exhibited cytoplasmic and nuclear localization. Thus, the loss of C-TAK1 binding has the same effect on Cdc25C as does mutation of the S216 phosphorylation site, indicating that C-TAK1 contributes to the regulation of S216 phosphorylation and Cdc25C intracellular localization in quiescent interphase cells.
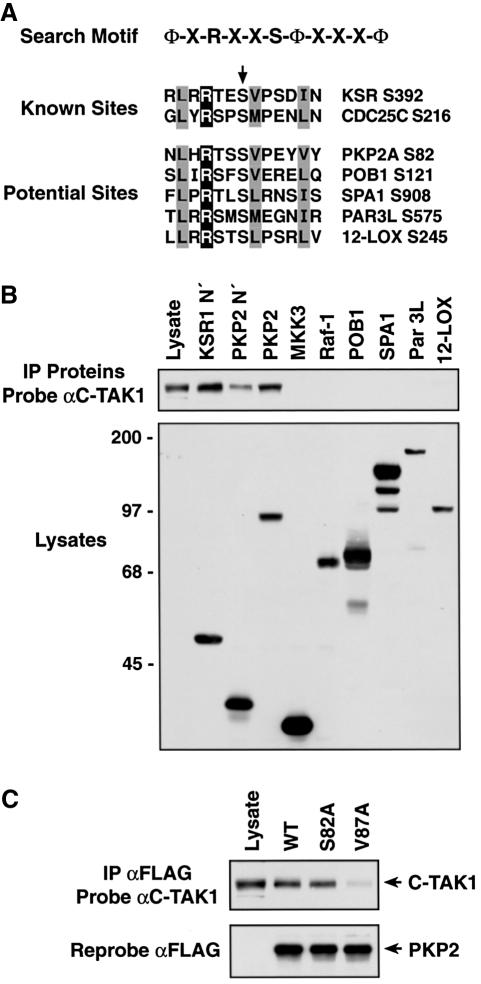
Identification of plakophilin 2 as a new C-TAK1 substrate
The above results suggest that the substrate recognition motif of C-TAK1 may be defined as ΦaxRxxS*ΦxxxΦa, where * is the site phosphorylated, x is any amino acid, Φa is a hydrophobic residue with aliphatic side chains and Φ is any hydrophobic amino acid. Moreover, our findings demonstrate that the hydrophobic residue at the +5 position is also required for the stable interaction of C-TAK1 with its targets KSR1 and Cdc25C. Therefore, to identify other potential in vivo substrates of C-TAK1, we searched the TrEMBL and Swiss-Prot databases for proteins that contain the following motif, ΦaxRxxSΦxxxΦa (Figure 6A). Expression constructs for several proteins identified in this search were obtained, including plakophilin 2 (PKP2), partitioning-defective 3-like protein (Par3L), platelet-type lipoxygenase 12 (LOXP), signal-induced proliferation-associated protein 1 (SPA1) and RalBP1-interacting protein (POB1). The proteins were then expressed in Cos cells and assessed for their ability to interact with C-TAK1 by co-immunoprecipitation assays (Figure 6B). KSR1 immune complexes served as a positive control for C-TAK1 binding, whereas immunoprecipitates of Raf-1 and MKK3 (proteins which do not contain the ΦaxRxxSΦxxxΦa motif) were included as negative controls. By immunoblot analysis, C-TAK1 was found to associate with KSR1 and both the full-length and N-terminal head domain (residues 1–348) of PKP2. As verification that C-TAK1 binding to PKP2 was mediated by the sequence identified in the motif search, we found that the interaction between C-TAK1 and PKP2 could be disrupted by mutation of the hydrophobic residue at the +5 position relative to the predicted phosphoacceptor site (V87A; Figure 6C).
Fig. 6. Binding of C-TAK1 to PKP2. (A) Sequences surrounding the known C-TAK1 phosphorylation sites of KSR1 (S392) and Cdc25C (S216) are shown. Also depicted are sequences from proteins containing the ΦaxRxxSΦxxxΦa motif, which were tested for C-TAK1 binding. (B) Cos cells were transfected with constructs encoding N′424 KSR1, POB1, Spa1, Par 3L, 12-LOX, MKK3, Raf-1 and either full-length or the N-terminal head domain of PKP2 (PKP2 N′). Proteins were immunoprecipitated from cell lysates, and the immune complexes were examined for the presence of C-TAK1. Lysates were probed with a mixture of antibodies recognizing the Pyo, FLAG, GFP, Myc and HA epitope tags to confirm the expression of the transfected proteins. (C) PKP2 proteins immunoprecipitated from Cos cells expressing wild-type, S82A or V87A PKP2 were examined by immunoblot analysis using C-TAK1 and FLAG antibodies.
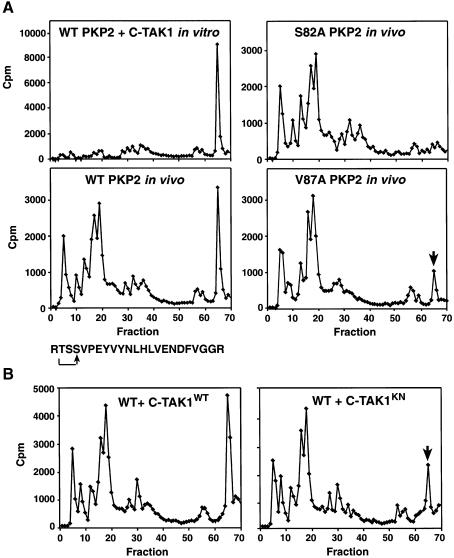
Next, to determine whether PKP2 is a substrate of C-TAK1, in vitro kinase assays were performed using affinity-purified PKP2 and C-TAK1. The phosphorylation state of PKP2 was then examined by HPLC analysis. As shown in Figure 7A, PKP2 was phosphorylated by C-TAK1 on a tryptic peptide that eluted in fraction 65 of the HPLC gradient. Through phosphoamino acid analysis and Edman degradation of the labeled peptide, the residue phosphorylated was determined to be S82 (the site predicted by the motif search). To investigate whether S82 is phosphorylated in intact cells and whether C-TAK1 contributes to the phosphorylation of S82, we examined the in vivo phosphorylation state of wild-type PKP2 and proteins mutated at either the S82 or V87 site (Figure 7A). By HPLC analysis, wild-type PKP2 contained a phosphopeptide eluting in fraction 65, which was not observed when a similar analysis was performed on the S82A mutant. In addition, phosphorylation of the fraction 65 peptide was reduced ∼72% for the PKP2 V87A mutant (Figure 7A) and was reduced ∼51% when wild-type PKP2 was expressed with kinase-negative C-TAK1 (Figure 7B). These findings identify S82 as a site of PKP2 phosphorylated in vivo and demonstrate that PKP2 is an authentic substrate of C-TAK1.
Fig. 7. PKP2 is a substrate of C-TAK1. (A) In vitro kinase assays were performed using purified C-TAK1 and purified PKP2 (top left panel). Wild-type, S82A and V87A PKP2 proteins expressed in Cos cells were labeled in vivo with [32P]orthophosphate (bottom left and right panels). The labeled PKP2 proteins were isolated, digested with trypsin and examined by HPLC analysis. Also shown is the sequence of the phosphorylated peptide eluting in fraction 65. (B) Quiescent Cos cells co-expressing wild-type PKP2 and either wild-type C-TAK1 or kinase-negative C-TAK1 (C-TAK1KN) were labeled in vivo with [32P]orthophosphate and analyzed as described in (A).
Phosphorylation of S82 mediates 14-3-3 binding and influences the subcellular localization of PKP2
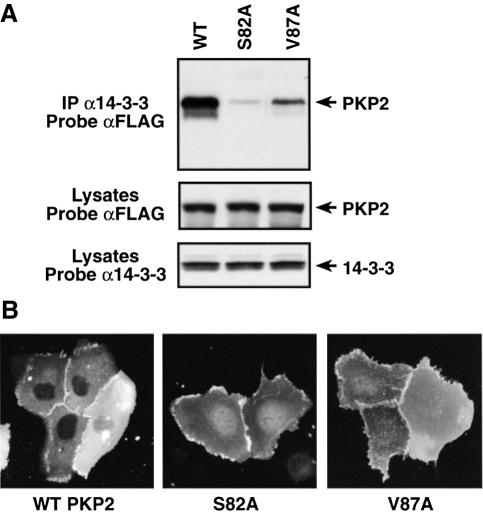
Given that the PKP2 S82 phosphorylation site is found within a canonical 14-3-3-binding motif (RSxS*xP; Yaffe et al., 1997), we next examined whether PKP2 interacts with 14-3-3. As shown in Figure 8A, PKP2 was found to co-immunoprecipitate with 14-3-3; however, this interaction could be disrupted by mutations that either destroy the S82 site (S82A) or severely reduce its phosphorylation (V87A), confirming that S82 is a functional 14-3-3-binding site.
Fig. 8. In vivo analysis of the PKP2–14-3-3 interaction. (A) Cos cells were transfected with constructs encoding 14-3-3 and wild-type, S82A or V87A PKP2. 14-3-3 proteins were immunoprecipitated from cell lysates, and the immune complexes were examined by immunoblot analysis using FLAG and 14-3-3 antibodies. (B) SCC-9 cells were transfected with constructs encoding wild-type, S82A or V87A PKP2. Localization of the PKP2 proteins was then determined by indirect immunofluorescence using the FLAG antibody.
To examine whether 14-3-3 binding influences the subcellular localization of PKP2, indirect immunofluorescent staining was performed on SCC-9 keratinocytes transiently expressing wild-type, S82A or V87A PKP2 (Figure 8B). PKP2 is an armadillo (ARM) repeat-containing protein that previously has been shown to localize in both desmosomes and the nucleus. As shown in Figure 8B, punctate staining at the cell border, which is characteristic of desmosomes, was observed in cells expressing wild-type PKP2. However, as reported by Chen et al. (2002), we found that wild-type PKP2 was rarely seen in the nucleus. While staining at the cell border was also observed in cells expressing either the S82A or V87A mutant, nuclear staining was considerably more pronounced than was observed in wild-type PKP2-expressing cells. The nuclear localization of the PKP2 mutants was confirmed by biochemical fractionation (data not shown). Thus, as with KSR1 and Cdc25C, the C-TAK1-mediated phosphorylation of a 14-3-3-binding motif contributes to the intracellular localization of PKP2.
Discussion
C-TAK1, a member of the CAMK group of kinases, has been shown to phosphorylate proteins on sites that generate 14-3-3-binding motifs. The two best characterized substrates of C-TAK1 are the Cdc25C phosphatase and the MAPK scaffold KSR1. Interestingly, not only does C-TAK1 phosphorylate these proteins, it has also been found to stably associate with them. In fact, C-TAK1 was first purified and cloned based on its interaction with Cdc25C, hence its name, Cdc25C-associated kinase 1. Subsequently, C-TAK1 was found to be a component of the KSR1 scaffolding complex. In this study, we have characterized the interaction of C-TAK1 with KSR1 and Cdc25C further and have identified key residues required for C-TAK1 binding and substrate phosphorylation. These findings have allowed us to screen protein databases, resulting in the identification of PKP2 as a new C-TAK1 substrate. We have extended this finding by showing that phosphorylation of a C-TAK1 consensus site in PKP2 generates a 14-3-3-binding site, which influences the intracellular localization of PKP2. These results demonstrate a conserved biochemical mechanism whereby C-TAK1 phosphorylation/14-3-3 binding contributes to the subcellular partitioning of proteins with diverse cellular functions.
Definition of the C-TAK1 substrate recognition motif
Insights into substrate binding and phosphorylation specificity by different members of the CAMK family have emerged from our analysis of C-TAK1 substrates. By comparing the sequences of Cdc25C and KSR1 known to contain the interaction site for C-TAK1 and subsequent mutational analysis of selected amino acids, we find that a hydrophobic residue at the +5 position relative to the site phosphorylated by C-TAK1 is essential for mediating the stable interaction between C-TAK1 and its substrates. Significantly, this residue is not only required for C-TAK1 binding, but is also critical for C-TAK1 substrate phosphorylation. Further mutational analysis of residues surrounding the C-TAK1 target phosphorylation site on KSR1 (S392) has revealed that an arginine residue at the –3 position as well as hydrophobic amino acids at the –5, +1 and +5 positions were required for efficient C-TAK1 phosphorylation. In terms of the relative importance for C-TAK1 substrate preference, we find that positions +5∼–3>–5>>+1.
Previous studies using synthetic peptide substrates or peptide library approaches to define phosphorylation recognition motifs have revealed that members of both the AGC and CAMK groups exhibit a preference for substrates that contain a basic residue at the –3 position relative to the phosphoacceptor site (Pearson and Kemp, 1991; Songyang et al., 1996). Further examination of these motifs indicates that while members of the AGC kinases tend to phosphorylate substrates that contain additional basic residues surrounding the phosphoacceptor site, members of the CAMK group often prefer targets that have hydrophobic residues in close proximity to the phosphoacceptor site. In particular, like C-TAK1, the CAMK members SNF1, AMPK, CAMKI, Chk1 and Chk2 have all been shown to have a strong preference for substrates containing hydrophobic residues at the –5 position and a weaker requirement for hydrophobic residues at the +1 position (Dale et al., 1995; Hutchins et al., 2000; O’Neill et al., 2002). In addition, similar to what we observe for C-TAK1, Chk1 and Chk2 prefer hydrophobic residues with aliphatic side chains at the –5 position (O’Neill et al., 2002). Residues C-terminal to the +1 site, however, have not been found to play a strong role in substrate recognition by the Chk1 and Chk2 kinases (Hutchins et al., 2000; O’Neill et al., 2002). In contrast, a hydrophobic residue at the +4 position relative to the phosphoacceptor site has been shown to be required for the AMPK, SNF1 and CAMKI kinases. Interestingly, substrate phosphorylation mediated by these kinases is abolished when the +4 hydrophobic residue is moved to the +3 or +5 position (Dale et al., 1995). Therefore, C-TAK1 appears to be unique in its requirement for a hydrophobic residue at the +5 position. Whether other EMK/MARK/Par1 kinases will have similar substrate recognition requirements awaits determination.
Identification of plakophilin 2 as a new C-TAK1 substrate
Using the information acquired from our mutational analysis examining C-TAK1 binding and substrate phosphorylation, a motif was defined (ΦaxRxxSΦxxxΦa) that was then used to search the TrEMBL and SwissProt databases for additional proteins that might be in vivo substrates of C-TAK1. Evaluation of proteins identified in this in silico search revealed that PKP2 is a C-TAK1-interacting protein. Although we did not examine all proteins that contain the ΦaxRxxSΦxxxΦa motif, of those tested, only PKP2 was found to interact with and be phosphorylated by C-TAK1. These findings suggest that either the ΦaxRxxSΦxxxΦa motif is inaccessible for binding and phosphorylation in the non-C-TAK1-interacting proteins or that there are additional determinants in the primary sequence or the folding of the domain that confers C-TAK1 binding. In support of the latter possibility, we have found recently that introduction of an arginine residue at the –1, +4 or +6 positions relative to the phosphoacceptor site greatly reduces the KSR1–C-TAK1 interaction, suggesting that the presence of basic amino acids in this region may impair C-TAK1 binding. Strikingly, all of the proteins that contain the ΦaxRxxSΦxxxΦa motif but did not interact with C-TAK1 have basic residues in the +2 to +6 positions.
In vivo analysis of C-TAK1-mediated phosphorylation
The generation of mutant substrate proteins that do not interact with C-TAK1 has provided us with the opportunity to evaluate the functional consequence of C-TAK1 binding in vivo. In particular, these mutants have allowed us to determine whether C-TAK1 is indeed a kinase that contributes to the phosphorylation state of KSR1 and Cdc25C in intact cells. Previously, KSR1 has been shown to be constitutively phosphorylated on two sites in resting cells, S297 and S392. Here, we find that disruption of C-TAK1 binding reduced the phosphorylation state of S392 ∼75%, while S297 phosphorylation was unaffected. Likewise, overexpression of a dominant-inhibitory C-TAK1 protein reduced phosphorylation of S392 but not S297. The specific reduction in S392 phosphorylation strongly indicates that C-TAK1 is the major kinase that phosphorylates this site in vivo. In addition, the finding that the phosphorylation state of S297 is not dependent on C-TAK1 binding, together with the observation that S297 is only weakly phosphorylated by purified C-TAK1 in vitro (Müller et al., 2001), suggests that the S297 site is phosphorylated by a different cellular kinase.
For Cdc25C, we found that the phosphorylation state of S216 was altered either by the loss of C-TAK1 binding or by overexpression of the dominant-inhibitory C-TAK1 protein, indicating that C-TAK1 contributes to the phosphorylation of this site in vivo. Interestingly, the S216 site of Cdc25C has also been found to be a substrate of Chk1 (Peng et al., 1997; Sanchez et al., 1997). Chk1 is a nuclear checkpoint kinase that plays an important role in preventing cell cycle progression during conditions of DNA damage. In this case, Chk1 becomes activated in response to DNA damage and phosphorylates nuclear-localized Cdc25C on S216, thus restoring 14-3-3 binding and inducing the export of Cdc25C from the nucleus. The fact that Cdc25C is a substrate for both C-TAK1 and Chk1 is consistent with the overlapping substrate phosphorylation requirements for Chk1 (ΦaxRxxS*Φ) and C-TAK1 (ΦaxRxxS*ΦxxxΦa). Adding further support to this idea, we found that mutation of the hydrophobic residue at the –5 position severely reduced S216 phosphorylation by either C-TAK1 or Chk1 in vitro, whereas mutation of the +5 hydrophobic amino acid only altered C-TAK1-mediated phosphorylation. Although these results might suggest that all C-TAK1 substrates should also be Chk1 substrates, we have found previously that the KSR1 S392 site is only weakly phosphorylated by purified Chk1 (Müller et al., 2001), indicating that there may be additional factors that influence substrate recognition.
For all three substrates analyzed in this study, KSR1, Cdc25C and PKP2, disrupting the binding of C-TAK1 had the same effect as did mutation of the site phosphorylated by C-TAK1. In particular, the subcellular localization of these proteins was affected in an equivalent manner by either alteration. The means by which C-TAK1 modulates the localization of its substrate proteins appears to be due to the fact that the residue phosphorylated by C-TAK1 serves as a 14-3-3-binding site. For KSR1, 14-3-3 binding to S392 regulates the translocation of KSR1 from the cytosol to the plasma membrane, while for Cdc25C, the binding of 14-3-3 to S216 regulates the nuclear–cytoplasmic shuttling of Cdc25C. Our finding that PKP2 mutated at the S82/14-3-3-binding site exhibits increased nuclear accumulation suggests that 14-3-3 binding is likely to be involved in the nuclear shuttling of PKP2 as well. Plakophilins, including PKP2, are a family of ARM repeat proteins that have been reported to localize in both desmosomes and the nucleus (Mertens et al., 1996; Schmidt et al., 1997; Klymkowsky, 1999). PKP2 is thought to be involved in desmosome assembly and organization; however, its function in the nucleus is unclear. Interestingly, when Chen et al. (2002) compared the localization of ectopically expressed PKP1 and PKP2, they found that PKP2 was much less efficient than PKP1 in localizing to the nucleus. Since the C-TAK1-generated 14-3-3-binding site is not conserved in the other PKP members, the reduced nuclear localization of PKP2 may reflect the ability of PKP2 to interact with 14-3-3. Whether the phosphorylation/dephosphorylation of PKP2 at S82 is regulated under various signaling conditions and whether 14-3-3 binding to this site has other effects on PKP2 function requires further investigation.
Concluding remarks
The results from this and previous studies suggest that both C-TAK1 and 14-3-3 are able to bind to the target protein at the same time. In support of this observation, structural studies analyzing 14-3-3 phosphopeptide complexes predict that the residue at the +5 position relative to the phosphoserine would be located outside of the 14-3-3-binding cleft (Yaffe et al., 1997). Therefore, the hydrophobic residue at this position would still be accessible to interact with C-TAK1. In addition, C-TAK1 appears to remain associated with its substrates even after the target site is phosphorylated and 14-3-3 is bound. The sustained interaction with its substrates, together with the close proximity of the residue required for C-TAK1 binding and the phosphoacceptor site, may have important functional consequences in terms of the regulation of C-TAK1 substrates. Specifically, these features may be key to maintaining the phosphorylation of the target site at a high level of stoichiometry. If, for example, 14-3-3 binding is disrupted and the site becomes dephosphorylated, the presence of C-TAK1 might immediately rephosphorylate the site and restore 14-3-3 binding. This property would allow C-TAK1 effectively to maintain 14-3-3 binding in quiescent cells, thereby preventing inappropriate MAPK signaling in the case of KSR1 or premature entry into mitosis in the case of Cdc25C. While further investigation is needed to determine how 14-3-3 binding is relieved under signaling conditions or entry into mitosis, our findings implicate C-TAK1 as an important regulator of protein localization events that have major consequences for the transmission of cellular signals as well as the maintenance of cellular quiescence.
Materials and methods
Antibodies and DNA constructs
The monoclonal antibody recognizing the Pyo-derived epitope tag has been reported previously (Therrien et al., 1996) and the C-TAK1 antibody was generated as described in Peng et al. (1998). Anti-FLAG and purified Chk1 were obtained from Upstate (Lake Placid, NY), anti-hemagglutinin (HA) from Roche (Indianapolis, IN) and anti-Myc (9E10) from Santa Cruz Biotechnology (Santa Cruz, CA). pcDNA3 constructs encoding wild-type KSR1 (residues 1–873) and N′424 (residues 1–424) have been described previously (Therrien et al., 1996). The pcDNA3-Pyo-Cdc25C construct was generated by standard PCR techniques using a human Cdc25C cDNA clone as template. Primers were designed such that two copies of the Pyo tag were added in-frame to the N-terminus of Cdc25C. To obtain the pcDNA3-Pyo-C-TAK1, full-length C-TAK1 was first cloned by RT–PCR using RNA from human 293 cells as template and the Superscript one-step RT–PCR kit from Invitrogen (Carlsbad, CA). The Pyo tag was then added in-frame to the N-terminus of C-TAK1. All constructs were verified by sequencing. pCMV5a-FLAG-PKP2a and pCMV5a-FLAG-PKP2a-H2 (Chen et al., 2002) were generously provided by Dr Kathleen Green (Northwestern University Medical School). pRSV-FLAG-MKK3 (from Dr John Blenis, Harvard Medical School), pK-Myc-Par3L (from Dr Ian Macara, University of Virginia Medical Center), pSK-SRα-Myc-Spa-1 (from Dr Nagahiro Minato, Kyoto University), pCGN-HA-POB1 (from Dr Akira Kikuchi, Hiroshima University School of Medicine) and pEGFP-C2/hP-12-LOX (from Dr Colin Funk, University of Pennsylvania Medical Center) were also used. Point mutations were introduced by site-directed mutagenesis (QuickChange; Stratagene, La Jolla, CA), and all mutations were confirmed by DNA sequencing.
Cell culture, transfection and co-immunoprecipitation assays
NIH-3T3 and Cos cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). SCC-9 cells were maintained in DMEM/Ham’s nutrient medium F12 (1:1 mixture) supplemented with 400 ng/ml hydrocortisone and 10% FBS. All cells were cultured at 37°C in 5% CO2. Plasmid DNAs were transfected into Cos and SCC-9 cells using the Fugene reagent (Roche), and into NIH-3T3 cells using Lipofectamine (Invitrogen). Transfected cells were either incubated for 48 h in fully supplemented medium or were serum starved for 18 h prior to analysis. For co-immunoprecipitation assays, cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed in NP-40 lysis buffer [20 mM Tris pH 8.0, 137 mM NaCl, 10% glycerol, 1% NP-40, 0.15 U/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride (PMSF), 20 µM leupeptin, 5 mM sodium vanadate] for 20 min at 4°C. Proteins were then immunoprecipitated from the cell lysates using the appropriate antibody and proteinA/G–Sepharose beads. After extensive washing in NP-40 lysis buffer, the immune complexes were examined by SDS–PAGE and immunoblotting.
In vitro kinase assays
Transfected Cos cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (NP-40 lysis buffer that contains 0.5% sodium deoxycholate and 0.1% SDS), and Pyo-tagged KSR1 or Cdc25C proteins were affinity purified as previously described (Müller et al., 2001). A 0.2 µg aliquot of purified protein was then incubated in kinase buffer [30 mM Tris pH 7.4, 1 mM dithiothreitol (DTT), 10 mM MnCl2, 1 µM ATP] containing 20 µCi of [γ-32P]ATP and either purified C-TAK1 or Chk1 for 30 min at room temperature. The assays were terminated by the addition of gel sample buffer (250 mM Tris pH 6.8, 50 mM DTT, 10% SDS, 30% glycerol).
Metabolic labeling and phosphorylation site mapping
Transfected Cos cells were incubated for 4–6 h at 37°C in phosphate-free DMEM containing 2.5% dialyzed calf serum and 1 mCi of [32P]orthophosphate per ml of labeling medium. Cells were then washed twice with ice-cold Tris-buffered saline (TBS) (20 mM Tris pH 7.4, 137 mM NaCl) and lysed in RIPA buffer. The labeled proteins were immunoprecipitated from the cell lysates and examined by SDS–PAGE. Labeled proteins were eluted from the gel matrix, digested with trypsin, and analyzed by reverse-phase HPLC (Cacace et al., 1999). HPLC fractions containing peaks of radioactivity were subjected to phospho amino acid analysis and semi-automated Edman degredation as previously described (Morrison et al., 1993).
Immunofluorescence and Xenopus oocyte maturation assay
NIH-3T3 and SCC-9 cells were seeded onto 18 mm glass coverslips, transfected with the appropriate cDNA constructs, and fixed 40–48 h following transfection. NIH-3T3 cells were fixed in a freshly prepared solution of 4% paraformaldehyde in PBS for 10 min at 25°C, following which they were permeabilized for 5 min in PBS containing 0.1% Triton X-100. SCC-9 cells were fixed and permeabilized by treatment of the cells with methanol for 2 min at –20°C (Chen et al., 2002). Both cell types were then washed with PBS and blocked in 3% bovine serum albumin (BSA) in PBS. Following incubation in the appropriate antibody, the cells were washed with PBS and incubated with either anti-mouse or anti-rabbit Alexa dye secondary antibody (Molecular Probes, Eugene, OR). The coverslips were then washed in PBS and mounted in Prolong antifade medium (Molecular Probes). The Xenopus oocyte maturation assays were performed as described in Müller et al. (2001).
Acknowledgments
Acknowledgements
We thank Drs Helen Piwnica-Worms, Kathleen Green, John Blenis, Ian Macara, Nagahiro Minato, Akira Kikuchi and Colin Funk for generously providing valuable constructs and reagents, and Dr Mark Fortini and members of the Morrison laboratory for helpful discussions and input.
References
- Brunet A. et al. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell, 96, 857–868. [DOI] [PubMed] [Google Scholar]
- Cacace A.M., Michaud,N.R., Therrien,M., Mathes,K., Copeland,T., Rubin,G.M. and Morrison,D.K. (1999) Identification of constitutive and ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding and KSR overexpression. Mol. Cell. Biol., 19, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Bonne,S., Hatzfeld,M., van Roy,F. and Green,K.J. (2002) Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and β-catenin signaling. J. Biol. Chem., 277, 10512–10522. [DOI] [PubMed] [Google Scholar]
- Dale S., Wilson,W.A., Edelman,A.M. and Hardie,D.G. (1995) Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1 and mammalian calmodulin-dependent protein kinase I. FEBS Lett., 361, 191–195. [DOI] [PubMed] [Google Scholar]
- Graves P.R., Lovly,C.M., Uy,G.L. and Piwnica-Worms,H. (2001) Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 protein binding. Oncogene, 20, 1839–1851. [DOI] [PubMed] [Google Scholar]
- Grozinger C.M. and Schreiber,S.L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA, 97, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins J.R., Hughes,M. and Clarke,P.R. (2000) Substrate specificity determinants of the checkpoint protein kinase Chk1. FEBS Lett., 466, 91–95. [DOI] [PubMed] [Google Scholar]
- Jaumot M. and Hancock,J.F. (2001) Protein phosphatases 1 and 2A promote Raf-1 activation by regulating 14-3-3 interactions. Oncogene, 20, 3949–3958. [DOI] [PubMed] [Google Scholar]
- Kao H.Y., Verdel,A., Tsai,C.C., Simon,C., Juguilon,H. and Khochbin,S. (2001) Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem., 276, 47496–47507. [DOI] [PubMed] [Google Scholar]
- Klymkowsky M.W. (1999) Plakophilin, armadillo repeats and nuclear localization. Microsc. Res. Tech., 45, 43–54. [DOI] [PubMed] [Google Scholar]
- Kubicek M., Pacher,M., Abraham,D., Podar,K., Eulitz,M. and Baccarini,M. (2002) Dephosphorylation of Ser-259 regulates Raf-1 membrane association. J. Biol. Chem., 277, 7913–7919. [DOI] [PubMed] [Google Scholar]
- Kumagai A. and Dunphy,W.G. (1999) Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev., 13, 1067–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light Y., Paterson,H. and Marais,R. (2002) 14-3-3 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane to maintain signaling fidelity. Mol. Cell. Biol., 22, 4984–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C.J. (1996) Ras effectors. Curr. Opin. Cell Biol., 2, 197–204. [DOI] [PubMed] [Google Scholar]
- Mertens C., Kuhn,C. and Franke,W.W. (1996) Plakophilins 2a and 2b: constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J. Cell Biol., 135, 1009–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.K. (2001) KSR: a MAPK scaffold of the Ras pathway? J. Cell Sci., 114, 1609–1612. [DOI] [PubMed] [Google Scholar]
- Morrison D.K., Heidecker,G., Rapp,U.R. and Copeland,T.D. (1993) Identification of the major phosphorylation sites of the Raf-1 kinase. J. Biol. Chem., 268, 17309–17316. [PubMed] [Google Scholar]
- Müller J., Ory,S., Copeland,T., Piwnica-Worms,H. and Morrison,D.K. (2001) C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell, 8, 983–993. [DOI] [PubMed] [Google Scholar]
- Muslin A.J. and Xing,H. (2000) 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal., 12, 703–709. [DOI] [PubMed] [Google Scholar]
- Muslin A.J., Tanner,J.W., Allen,P.M. and Shaw,A.S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell, 84, 889–897. [DOI] [PubMed] [Google Scholar]
- Ogg S., Gabrielli,B. and Piwnica-Worms,H. (1994) Purification of a serine kinase that associates with and phosphorylates human Cdc25C on serine 216. J. Biol. Chem., 269, 30461–30469. [PubMed] [Google Scholar]
- O’Neill T. et al. (2002) Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J. Biol. Chem., 277, 16102–16115. [DOI] [PubMed] [Google Scholar]
- Pearson R.B. and Kemp,B.E. (1991) Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol., 200, 62–81. [DOI] [PubMed] [Google Scholar]
- Peng C.-Y., Graves,P.R., Thomas,R.S., Wu,Z., Shaw,A.S. and Piwnica-Worms,H. (1997) Miotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25c on serine 216. Science, 277, 1501–1505. [DOI] [PubMed] [Google Scholar]
- Peng C.Y., Graves,P.R., Ogg,S., Thoma,R.S., Byrnes,M.J., Wu,Z., Stephenson,M.T. and Piwnica-Worms,H. (1998) C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ., 9, 197–208. [PubMed] [Google Scholar]
- Sanchez Y., Wong,C., Thoma,R., Richman,R., Wu,Z., Piwnica-Worms,H. and Elledge,S. (1997) Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regultion through Cdc25. Science, 277, 1497–1501. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Langbein,L., Rode,M., Pratzel,S., Zimbelmann,R. and Franke,W.W. (1997) Plakophilins 1a and 1b: widespread nuclear proteins recruited in specific epithelial cells as desmosomal plaque components. Cell Tissue Res., 290, 481–499. [DOI] [PubMed] [Google Scholar]
- Songyang Z. et al. (1996) A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5 and Erk1. Mol. Cell. Biol., 16, 6486–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.Q., Lu,B., Feng,J.J., Reinhard,C., Jan,Y.N., Fantl,W.J. and Williams,L.T. (2001) PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat. Cell Biol., 3, 628–636. [DOI] [PubMed] [Google Scholar]
- Therrien M., Michaud,N.R., Rubin,G.M. and Morrison,D.K. (1996) KSR modulates signal propagation within the MAPK cascade. Genes Dev., 10, 2684–2695. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B. (2002a) How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett., 513, 53–57. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B. (2002b) Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell. Biol., 3, 177–186. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B., Rittinger,K., Volinia,S., Caron,P.R., Aitken,A., Leffers,H., Gamblin,S.J., Smerdon,S.J. and Cantley,L.C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]
- Yang J., Winkler,K., Yoshida,M. and Kornbluth,S. (1999) Maintenance of G2 arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J., 18, 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.H., Kobayashi,R., Graves,P.R., Piwnica-Worms,H. and Tonks,N.K. (1997) Serine phosphorylation-dependent association of the band 4.1-related protein-tyrosine phosphatase PTPH1 with 14-3-3β protein. J. Biol. Chem., 272, 27281–27287. [DOI] [PubMed] [Google Scholar]
- Zhou M., Horita,D.A., Waugh,D.S., Byrd,R.A. and Morrison,D.K. (2002) Solution structure and functional analysis of the cystein-rich C1 domain of kinase suppressor of ras (KSR). J. Mol. Biol., 315, 435–446. [DOI] [PubMed] [Google Scholar]