Abstract
The dental follicle, a loose connective tissue sac that surrounds the unerupted tooth, appears to regulate the osteoclastogenesis needed for eruption; i.e., bone resorption to form an eruption pathway. Thus, DNA microarray studies were conducted to determine which chemokines and their receptors were expressed chronologically in the dental follicle, chemokines that might attract osteoclast precursors. In the rat first mandibular molar, a major burst of osteoclastogenesis occurs at day 3 with a minor burst at day 10. The results of the microarray confirmed our previous studies showing the gene expression of molecules such as CSF-1 and MCP-1 in the dental follicle cells. Other new genes also were detected, including secreted frizzled-related protein-1 (SFRP-1), which was found to be down-regulated at days 3 and 9. Using rat bone marrow cultures to conduct in vitro osteoclastogenic assays, it was demonstrated that SFRP-1 inhibited osteoclast formation in a concentration-dependent fashion. However, with increasing concentrations of SFRP-1, the number of TRAP-positive mononuclear cells increased suggesting that SFRP-1 inhibits osteoclast formation by inhibiting the fusion of mononuclear cells (osteoclast precursors). Co-culturing bone marrow mononuclear cells and dental follicle cells demonstrated that the dental follicle cells were secreting a product(s) that inhibited osteoclastogenesis, as measured by counting of TRAP-positive osteoclasts. Adding an antibody either to SFRP-1 or OPG partially restored osteoclastogenesis. Adding both anti-SFRP-1 and anti-OPG fully negated the inhibitory effect of the follicle cells upon osteoclastogenesis. These results strongly suggest that SFRP-1 and OPG, both secreted by the dental follicle cells, use different pathways to exert their inhibitory effect on osteoclastogenesis. Based on these in vitro studies of osteoclastogenesis, it is likely that the down-regulation of SFRP-1 gene expression in the dental follicle at days 3 and 9 is a contributory factor in allowing the major and minor bursts of osteoclastogenesis to occur. Thus, inhibition of SFRP-1 gene expression in combination with inhibition of OPG gene expression likely are critical events in enabling alveolar bone resorption to occur such that teeth will erupt.
Keywords: Dental follicle, Microarray, Osteoclastogenesis, SFRP-1, Tooth eruption
Introduction
Osteoclastogenesis and subsequent bone resorption are major requirements for tooth eruption [see review by 1]. To accomplish this requires the presence of the dental follicle, a loose connective tissue sac that surrounds the unerupted tooth. Specifically, mononuclear cells (osteoclast precursors) are recruited to the follicle [2, 3] where they fuse to form osteoclasts for alveolar bone resorption of the eruption pathway. In the first mandibular molar of the rat, the maximal number of mononuclear cells and osteoclasts are seen at day 3 [3, 4] and this correlates with the maximal gene expression of 2 chemotactic molecules in the follicle – colony stimulating factor-1 (CSF-1) [5] and monocyte chemotactic protein-1 (MCP-1) [6]. These molecules are chemotactic for monocytes in vitro and they are secreted and chemotactically active by the dental follicle [6].
Prior to the time of eruption of the 1st mandibular molar in the rat at day 18 postnatally, there is a major burst of osteoclastogenesis at day 3 [3, 4] and a minor burst at day 10 [4]. A ratio of receptor activator of nuclear factor kappa B ligand (RANKL) to osteoprotegerin (OPG) that reflects more RANKL will promote osteoclastogenesis [e.g., see review by 7]. Thus, for the major burst of osteoclastogenesis at day 3 we have shown that the gene expression of OPG in the follicle is downregulated by the CSF-1 produced in the follicle to promote a favorable ratio for osteoclastogenesis [8]. At day 10, the expression of OPG in the follicle is higher because of the reduced expression of CSF-1 but the gene expression of RANKL in the follicle is elevated [9]. Consequently, a slightly favorable ratio of RANKL/OPG likely is established to allow a minor burst of osteoclastogenesis.
Based on the above, the first aim of this study was to determine if other molecules might be involved in the recruitment of osteoclast precursors and/or the subsequent osteoclastogenesis needed for tooth eruption. DNA microarray studies were conducted to determine which chemokines and their receptors were expressed chronologically in the dental follicle in vivo. These studies suggested that another gene, secreted frizzled-related protein-1 (SFRP-1) had a reduced expression at both day 3 and day 9; i.e., times that correspond to the osteoclastogenic bursts. Thus, the second aim of this study was to determine the role of SFRP-1 in osteoclastogenesis, as determined by in vitro osteoclastogenesis assays.
Materials and methods
Dental follicle isolation and cell culture
Dental follicles were surgically isolated from the first mandibular molars of rats (Harlan Sprague-Dawley) at postnatal days 1, 3, 5, 7, 9 and 11 for RNA isolation. For cell culture, the dental follicles of the first mandibular molar were isolated from 5–6 day old rats and trypsinized to obtain the dental follicle cells, as previously described [10]. The dental follicle cells were cultured in minimum essential medium (MEM) (Sigma-Aldrich, St Louis, MO, USA) plus 10% (v/v) newborn calf serum, 1 mM sodium pyruvate, 1% penicillin/streptomycin and 0.2% fungazone, in an incubator at 37 °C with 5% CO2 until passage 5–6. Follicle cells of passages 6–9 were used for experiments.
RNA isolation
Total RNA was isolated from the dental follicles or the dental follicle cells using Tri-Reagent (Molecular Research Center, Cincinnati, OH, USA). The RNA then was treated with DNase I (Ambion, Austin, TX, USA) to remove DNA contaminations. For the microarray study, RNA was further purified using RNeasy® mini kit (Qaigen Inc., Valencia, CA, USA) to obtain highly pure RNA. Total RNA was quantified by a spectrophotometer at an absorbance (A) of 260nm, and RNA purity was confirmed by an A260: A280 ratio of 2.0 or higher.
Oligo DNA microarray analysis
Purified RNA (1–1.5 μg) was labeled with biotin-16-UTP (Roche Applied Science, Indianapolis, IN, USA) to generate complementary RNA (cRNA) using TrueLabeling-AMP™ 2.0 kit (SuperArray Bioscience, Frederick, MD, USA) per manufacturer’s instructions. The labeled cRNA was purified using RNeasy® Mini Kit (Qaigen Inc.) and quantified by a spectrophotometer. Four (4) μg of the labeled cRNA was hybridized to a rat chemokines & receptors oligo DNA microarray that contained 113 chemokines and receptors genes (SuperArray Bioscience, Cat. No. ORN-022). The hybridization was performed in the hybridization tube with 0.75ml hybridization buffer which was placed in a hybridization oven at 60°C for 18 h with gentle agitation (5–10 rpm), followed by the first wash in 2 × SSC, 1 % SDS at 60°C for 15 min, and the second wash in 0.1 × SSC, 0.5 % SDS at 60°C for 10 min. After a second wash, the array was blocked in blocking buffer at room temperature for 40 min, incubated with alkaline phosphatase-conjugated streptavidin for 10 min, and washed with wash buffer four times, 5 minutes each. Finally, the array was incubated at room temperature with a CDP-Star chemiluminescent substrate for 5 min.
The image was acquired with 20 minutes of exposure using FluoChem™ 8800 image system (Alpha Innotech Corp., San Leandro, CA, USA). The gene expression data were obtained and analyzed using software GEArray Expression Analysis Suite (SuperArray Bioscience), and gene expression level was expressed as the ratio of a given gene to the control glyceraldehyde-3-phosphate dehydrogenase (GAPD) gene.
For the in vivo studies, three independent litters of rats were used. For the in vitro studies, the experiments were repeated three times.
Reverse transcription polymerase chain reaction (RT-PCR)
To detect gene expression, 2 μg total RNA was reverse transcribed by M-MLV Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) to synthesize the first strand cDNA in a 20 μl reaction volume per manufacturer’s instructions. The reverse transcription reaction was performed at 37°C for 1 h, and then incubated at 70°C for 10 min to inactivate the activity of reverse transcriptase.
To confirm the expression of secreted frizzled-related protein-1 (SFRP-1) gene in the dental follicle cells, forward primer 5′-TAAAGAATGGCGCCGACTGTC-3′ and reverse primer 5′-TGGCTGTGAGCAAGTACTGGCT-3′ were designed based on partial mRNA sequence of the rat SFRP-1 gene (Gen Bank accession no.: AF167308). PCR was performed in a 25 μl reaction volume containing 2 μl of first strand cDNA as template, 1 × PCR reaction buffer, 200 μM each dNTPs, 300 nM each primer, and 1.5 units of Taq DNA polymerase. The reaction was carried out in a PTC-100 Programmable Thermal Controller for 28 cycles, with denaturation at 94°C for 45 s, annealing at 58°C for 50 s, and extension at 72°C for 55 s. After the PCR reaction, 10 μl of PCR product was run on agarose gel and stained with SYBR green I dye.
Immunostaining
For detection of SFRP-1 protein expression in vivo, mandibles of postnatal rats of day 5 were fixed in formalin, decalcified, and dehydrated in a series of graded alcohols. The mandibles were then embedded in paraffin and sectioned at 5 μm thickness. After de-paraffinization and rehydration, endogenous peroxidase activity was quenched by incubating the sections in 3% hydrogen peroxide for 10 min, followed by two washes with Tris-buffered saline (TBS) plus 0.025% Triton X-100, five minutes each. The sections were blocked for 1 h at room temperature with 1% BSA and 2% normal goat serum (Vector Laboratories, Burlingame, CA) in TBS buffer, and then incubated with rabbit polyclonal anti-human SFRP-1 primary antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA), which cross reacts with rat SFRP-1, at a concentration of 2 μg/ml overnight at 4 °C in TBS buffer plus 1% BSA and 2% normal goat serum. For controls, the primary antibody was omitted from the procedure. After three washes with TBS buffer plus 0.025% Triton X-100, the sections were incubated with biotinylated goat anti-rabbit IgG secondary antibody (1:100 dilutions in TBS buffer plus 1% BSA) for 1h. Following three washes, the sections were incubated with avidin-biotinylated horseradish peroxdase (HRP) (Vector Laboratories) for 30 min. The sections were incubated with NovaRED™ substrate (Vector Laboratories) for 5 min, followed by counterstaining with hematoxylin.
In vitro osteoclastogenesis and TRAP staining
Bone marrow cells were obtained from tibiae and femora of about six-week old rats by flushing the bone cavity with 15 ml α-MEM medium (Invitrogen) plus 10% (v/v) heated-inactivated fetal calf serum (FCS), 1% penicillin/streptomycin and 0.2% fungizone. After centrifugation at 1000 rpm for 5 min at room temperature, the cell pellet was re-suspended into 5 ml red blood cell lysing buffer (Sigma-Aldrich Corp.) for 5 min, and the cell suspension was centrifuged again under the same condition. The cell pellet was re-suspended in 5 ml PBS buffer, and the resulting cell suspension was loaded onto 5 ml Histopaque®-1077 (Sigma-Aldrich Corp.), followed by density gradient centrifugation at 2000 rpm for 30 min. The interface cell layer was harvested, washed with PBS buffer and centrifuged at 1000 rpm for 5 min to obtain mononuclear cells.
The mononuclear cells were cultured in a T-75 flask with 25 ml α-MEM medium plus 5 ng/ml human CSF-1 (Pepro Tech, Rock Hill, NJ, USA) for 24 h to allow stromal cells to adhere. After 24 h incubation, non-adherent cells were centrifuged at 1000 rpm for 5 min, and the resultant stromal-free bone marrow mononuclear cells were re-suspended in the same α-MEM medium as above at a density of approximately 2 × 105 cells per well in a 12-well plate with cytokines added in the following treatments: (1) CSF-1 only (75 ng/ml); (2) CSF-1 (75 ng/ml) and mouse RANKL (30 ng/ml) (R & D Systems); (3) CSF-1, RANKL, and human SFRP-1 (100 ng/ml) (R & D Systems); (4) CSF-1, RANKL, SFRP-1 (500 ng/ml); (5) CSF-1, RANKL, SFRP-1 (1000 ng/ml); (6) CSF-1, RANKL, SFRP-1 (500 ng/ml ) and additional RANKL (50 ng/ml). The cultures were placed in an incubator at 37°C with 5% CO2 for 5–6 days with the medium and cytokines changed every other day. The experiments were repeated three times.
For TRAP staining, cells were fixed and stained using a Leukocyte Acid Phosphatase kit according to manufacturer’s instructions (Sigma-Aldrich Corp.). The TRAP-positive (TRAP+) multinucleated osteoclasts and mononuclear cells per well (4 cm2) were counted separately under an inverted microscope at 100 X. The experiments were repeated three times.
Co-culture of bone marrow cells and dental follicle cells
Co-culture of bone marrow cells (BMC) and dental follicle cells (DFC) was performed in Costar Transwell® 24-well plates (Costar Corning, Cambridge, MA, USA). One day before co-culture, the dental follicle cells (DFC) in 100 μl MEM medium were cultured in an upper chamber which had a polycarbonate membrane with 0.4 μm pores to separate the DFC and BMC; i.e., the DFC did not contact the BMC. For co-culture studies, bone marrow mononuclear cells were added to 600 μl α-MEM medium in the lower chamber at a density of approximately 1 × 105 in the presence of 30 ng/ml CSF-1 and 10 ng/ml RANKL for the following treatments: (1) absence of DFC as a control; (2) DFC only; (3) DFC plus 500 ng/ml anti-SFRP-1 (R & D Systems); and (4) DFC plus 200 ng/ml anti-OPG (R & D Systems); (5) DFC plus 500 ng/ml anti-SFRP-1 and 200 ng/ml anti-OPG. Fresh MEM medium was added in the upper chamber. The cells were co-cultured at 37°C with 5% CO2, with medium, cytokines and antibody changed every day. After 7 days of co-culture, cells in the lower chamber were stained for TRAP+ osteoclasts, and the TRAP+ multinucleated osteoclasts per well (2 cm2) were counted. The co-culture experiments were repeated four times.
Statistical analysis
Data from the in vivo DNA microarray and TRAP-positive cell counting were analyzed using SAS program (version 9). Analysis of variance (ANOVA) was carried out to evaluate either the chronological changes of SFRP-1 gene expression or the effects of treatments on TRAP-positive cell numbers. The means were separated with the least significant difference (LSD) test at a significant level of P ≤ 0.05. The data were reported as mean ± standard error of three or four replications.
Results
Gene expression profiling in dental follicle cells
In the in vitro microarray studies, the expression in the follicle cells of several genes previously shown to have a role in recruitment of mononuclear cells and regulation of osteoclastogenesis for eruption was detected. These included colony-stimulating factor-1 (CSF-1), monocyte chemotactic protein-1 (MCP-1) and nuclear factor kappa-B1 (NFkB1). Additional genes included monocyte chemotactic protein-3 (MCP-3), melanoma growth stimulating activator1 (Cxcl1), myeloid differentiation primary response gene 88 (Myd88), chemokine orphan receptor 1 (Cmkor1), endothelial monocyte-activating polypeptide 2 (EMAP-II), secreted frizzled-related protein-1 (SFRP-1), peptidylprolyl isomerase A (Ppia), type 1 tumor necrosis factor receptor shedding aminopeptidase regulator (Arts1), chemokine-like factor 1 (Cklf1), colony stimulating factor 2 (CSF-2), matrix metallopeptidase 2 (Mmp2), cytoplasmic FMR1 interacting protein 2 (Cyfip2), stomal cell derived factor 2 (SDF-2), stromal cell-derived factor 2-like 1 (Sdf2l1), chemokine (C-C motif) ligand 11 (Ccl11), regulator of G-protein signaling 3 (RGS3), tumor necrosis factor receptor superfamily member 1a (Tnfrsf1a), ribosomal protein L32 (Rpl32), lactate dehydrogenase A (Ldha), and aldolase A (Aldoa) (Fig. 1A).
Fig. 1.
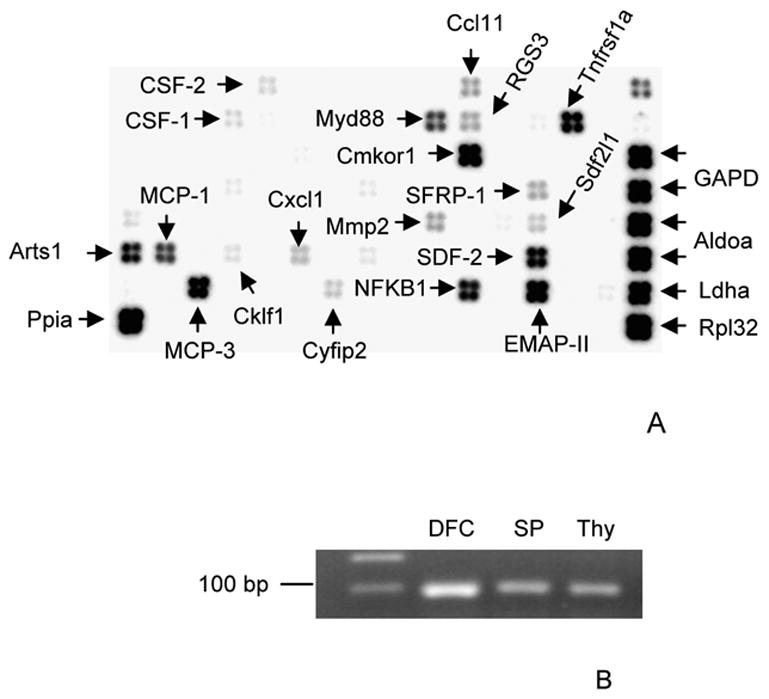
A) Arrows mark the expression of SFRP-1, CSF-1, MCP-1, NFkB1, MCP-3, Cxcl1, Myd88, Cmkor1, and EMAP-II in the dental follicle cells seen by the oligo DNA microarray. In addition to these putative tooth eruption genes, another 14 genes are seen in the microarray. B) The SFRP-1 gene expression identified in microarray was confirmed in the dental follicle cells (DFC) using RT-PCR. The RNAs of spleen (SP) and thymus (Thy) were also included in the RT-PCR as positive controls.
SFRP-1 (Fig. 1A) was chosen for further study because of the report that SFRP-1 was an inhibitor of osteoclastogenesis in mice [11]. To confirm the expression of the SFRP-1 gene in the rat follicle cells, RT-PCR was conducted using primers based on the partial gene sequence (Genbank accession no.: AF167368). As seen in Figure 1B, SFRP-1 was expressed.
Chronological expression of secreted frizzled-related protein-1 gene in dental follicle
In vivo expression of SFRP-1 in the dental follicle was examined using the chemokines & receptors DNA microarray, as in the in vitro study. The results showed that SFRP-1 was expressed in the dental follicle at days 1, 3, 5, 7, 9 and 11 (Fig. 2A), but expression levels at days 3 and 9 were lower than other days as determined by the ratio of SFRP-1 to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene. The lower expression of SFRP-1 at day 3 and day 9 was statistically significant as compared to that at 1, 5, 7 and 11 (Fig. 2B).
Fig. 2.
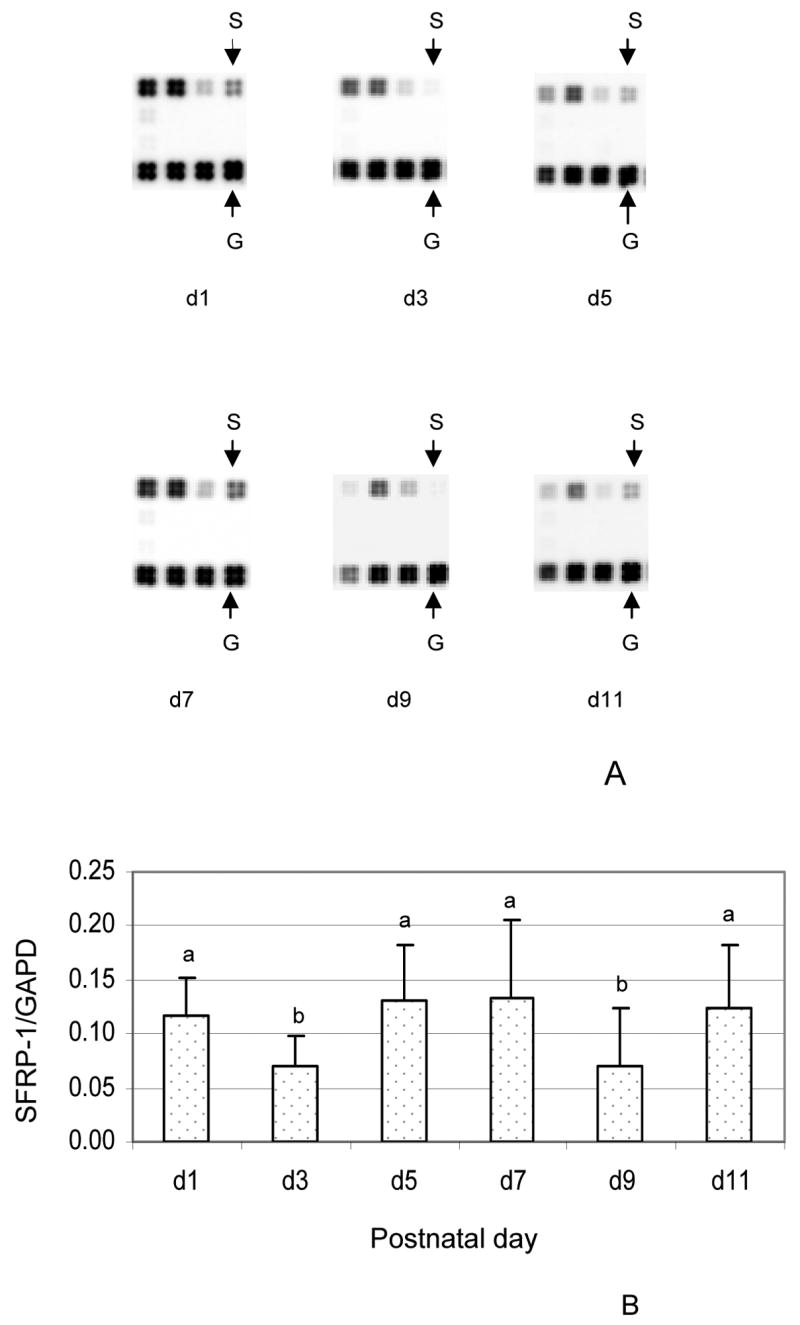
A) In vivo expression of secreted frizzled-related protein 1 (SFRP-1) gene in the dental follicle of rat as determined by the oligo DNA microarray for postnatal days 1, 3, 5, 7, 9 and 11. The images of the SFRP-1 gene (S) and internal control glyceraldehyde-3-phosphate dehydrogenase gene (G) are shown at the different days. B) Gene expression was expressed as the ratio of SFRP-1 gene to the internal control glyceraldehyde-3-phosphate dehydrogenase (GAPD) gene. The results were presented as means ± standard deviations of three independent litters of rats. The bars labeled with “b” are significantly different from those labeled with “a”. Note that the expression of SFRP-1 was lower at day 3 and day 9 in comparison to the other days.
In vivo expression of secreted frizzled-related protein-1 protein in dental follicle
To determine if SFRP-1 mRNA was translated in the dental follicle, mandible sections of rats at day 5 were immunostained for protein expression. The dental follicle strongly stained for SFRP-1 protein (Fig. 3), but there was an absence of staining in the stellate reticulum (Fig. 3). Some staining of the apex of ameloblasts stained, which likely represents a physical entrapment of the antibody. Slight staining of the alveolar bone also was observed.
Fig. 3.
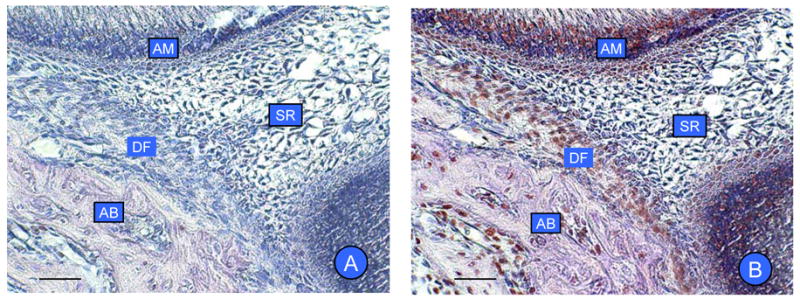
In vivo expression of SFRP-1 protein by immunohistochemistry in rat mandibular molars. (A) Controls for immunostaining in the absence of the primary antibody showing no staining. (B) Expression of SFRP-1 is seen in the dental follicle (DF). There is expression in ameloblasts (AM) and alveolar bone (AB), but no expression in stellate reticulum (SR). Scale bar: 50 μm.
Inhibition of osteoclastogenesis in bone marrow cell culture by secreted frizzled-related protein-1
To determine if SFRP-1 inhibits osteoclastogenesis in the rat, stromal cell-free bone marrow mononuclear cells were cultured in the presence of CSF-1 and RANKL, plus different concentrations of human SFRP-1, followed by TRAP staining and TRAP-positive (TRAP+) cell counting. The results showed that SFRP-1 inhibited osteoclastogenesis in a concentration-dependent manner, resulting in reduction of osteoclasts both in size and number. As shown in Fig. 4 and Table 1, TRAP+ osteoclasts (TRAP+ multinucleated cells) were formed in the culture without SFRP-1, with more than 50 nuclei in some giant cells (Fig. 4B). Addition of 100 ng/ml SFRP-1 to the culture slightly reduced the osteoclast number (Fig. 4C and Table 1). As the SFRP-1 concentration was increased to 500 ng/ml, the size and number of TRAP+ osteoclasts were reduced greatly (Fig. 4D and Table 1). At a concentration of 1000 ng/ml SFRP-1, almost all of the TRAP+ osteoclasts were small, typically with 2 to 5 nuclei, and the osteoclast number was reduced by more than 60% as compared with cultures without SFRP-1 (Fig. 4E and Table 1). Concomitant with the decrease of TRAP+ osteoclasts in size and number, the number of TRAP+ mononuclear cells was increased (Table 1). The inhibitory effect of SFRP-1 at concentration of 500 ng/ml can be negated by additional RANKL at a concentration of 50 ng/ml (Fig. 4F and Table 1).
Fig. 4.
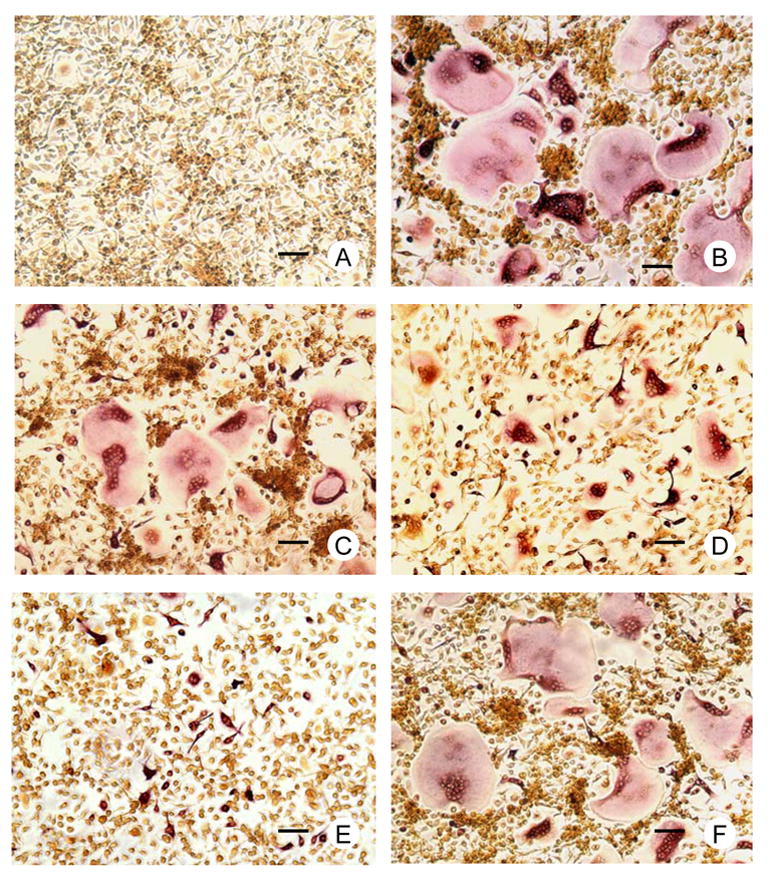
Effect of SFRP-1 on osteoclastogenesis in rat bone marrow cell cultures. The bone marrow mononuclear cells were cultured in the presence of various combinations of CSF-1, RANKL and SFRP-1. TRAP staining was performed and the TRAP-positive cells per well were counted. (Fig. 4A), no osteoclasts (TRAP+ multinucleated cells) are formed in cultures with CSF-1 only; (4B), larger osteoclasts are formed in culture with CSF-1 and RANKL; (4C, D and E), in the presence of CSF1 and RANKL, addition of 100 ng/ml (C), 500 ng/ml (D) and 1000 ng/ml (E) SFRP-1 to the cultures respectively resulted in smaller and fewer osteoclasts with a maximal reduction in numbers using 1000 ng/ml of SFRP-1; (4F), in the presence CSF-1, RANKL and 500 ng/ml SFRP-1, addition of another 50 ng/ml RANKL restored the osteoclast size and numbers. Scale bar: 50 μm.
Table 1.
Effect of SFRP-1 on number of osteoclasts and TRAP-positive mononuclear cells*
| Treatments (ng/ml) | Osteoclasts** | Mononuclear cells** |
|---|---|---|
| CSF-1 (75)+RANKL(30) | 2068 ± 370 a | 299 ± 114 B |
| CSF-1(75)+RANKL(30)+SFRP-1(100) | 1741 ± 255 b | 536 ± 180 B |
| CSF-1(75)+RANKL(30)+SFRP-1(500) | 1320 ± 172 c | 827 ± 205 B |
| CSF-1(75)+RANKL(30)+SFRP-1(1000) | 868 ± 65 d | 1593 ± 751 A |
| CSF-1(75)+RANKL(30)+SFRP-1(500)+RANKL(50) | 1966 ± 358 ab | 678 ± 297 B |
Based on cell counting per well in 12-well plate (4 cm2).
Cell numbers followed by different letter (s) within a column indicated a significant difference at P≤0.05 with the LSD test. For example, under “osteoclasts” “a” differs from all others under the column except for “ab”.
Inhibitory effect of osteoclastogenesis by dental follicle cells was partially reversed by anti-secreted frizzled-related protein-1
To determine if the dental follicle cells inhibit osteoclastogenesis by expression of SFRP-1, bone marrow cells were co-cultured with the dental follicle cells in the presence of CSF-1, RANKL and anti-SFRP-1. Because OPG also is secreted by the dental follicle cells and inhibits osteoclastogenesis, anti-OPG was included in the co-culture experiments for comparison. As shown in Fig. 5, co-culture of dental follicle cells resulted in reduction of osteoclastogenesis by about 60% in osteoclast number. Adding either anti-SFRP-1 or anti-OPG to the co-culture partially reduced the inhibitory effects, significantly increasing the osteoclast number. Adding both anti-SFRP-1 and anti-OPG to the co-culture almost completely neutralized the inhibitory effect by dental follicle cells; i.e., osteoclast numbers were restored to the level seen in the absence of the dental follicle cells.
Fig. 5.
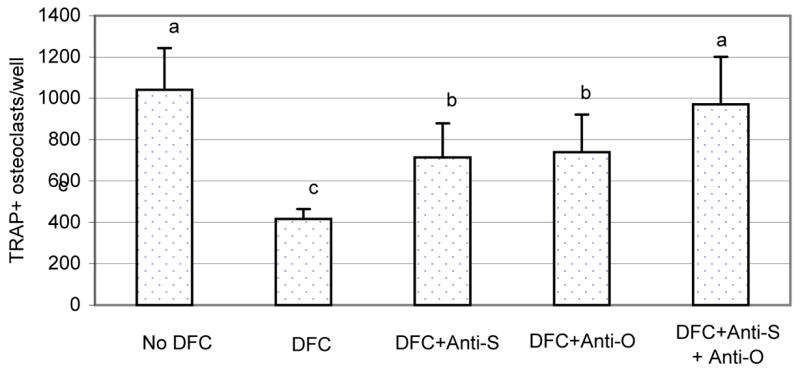
The bone marrow cells were co-cultured with the dental follicle cells in the presence of CSF-1 and RANKL, as well as anti-SFRP-1 (Anti-S) and anti-OPG (Anti-O) for 7 days. The bone marrow cell culture was then stained for TRAP+ osteoclasts, and TRAP+ osteoclasts per well were counted. Note that the presence of the dental follicle cells inhibited osteoclastogenesis but addition of either anti-SFRP-1 or anti-OPG in the culture increased osteoclast numbers. Addition of both antibodies essentially negated the inhibitory effect of the dental follicle cells. The bars with different letters between them indicate a statistically significant difference.
Discussion
DNA microarray has become a powerful tool for gene discovery and gene expression, enabling the analysis of many genes at the same time [12]. Using DNA microarray technology, the expression of CSF-1, MCP-1 and NFKB1 in the dental follicle was found to be maximal at day 3, confirming our previous studies [5, 6, 13]. At the same time, several other genes such as MCP-3, Cxcl1, Myd88, Cmkor1, EMAP-II and SFRP-1, for the first time were found to be expressed in the dental follicle. This expression profiling revealed by DNA microarray is consistent with results obtained by other methods such as RT-PCR.
One gene (SFRP-1) detected by this microarray study was of interest because of its chronological expression in the dental follicle and its possible function in osteoclastogenesis. SFRP-1 is a secreted membrane protein that binds directly to Wnt proteins, thereby inhibiting the Wnt signaling pathway [14]. Because of its ability to inhibit osteoclast formation in vitro, it was identified as an osteoclastogenesis inhibitor that was expressed by osteoblasts and chondrocytes in murine bone and cartilage [11]. Its inhibitory role in osteoclastogenesis also was demonstrated in studies in which bone marrow cells isolated from SFRP-1 knockout mice differentiated into osteoclasts 3- to 50-fold more than such cells from wild-type mice [15]. Our study indicated that the SFRP-1 gene was expressed in dental follicle cells, as well as it and the SFRP-1 protein being expressed in vivo in the dental follicle. The gene expression of SFRP-1 in the dental follicle, an essential connective tissue sac that regulates bone resorption and bone formation required for tooth eruption, was significantly lower at postnatal days 3 and 9, as compared to other days. Day 3 and day 10 are the times the major and minor bursts of osteoclastogenesis respectively occur [3, 4]. This led us to speculate that SFRP-1 may inhibit osteoclastogenesis, as seen in mice [11, 15]. This was confirmed by our in vitro osteoclastogenesis studies, showing that SFRP-1 inhibited osteoclastogenesis in vitro (Fig. 4). Moreover, cultures of dental follicle cells can secret SFRP-1 such that when they are co-cultured with osteoclast precursors (bone marrow cells), osteoclastogenesis is reduced (Fig. 5).
Our study further suggested that SFRP-1 may inhibit osteoclastogenesis by inhibiting fusion of osteoclast precursors (mononuclear cells). In bone marrow cell cultures, increasing SFRP-1 concentrations reduced both the size and number of osteoclasts but the number of TRAP-positive mononuclear cells were increased. This strongly suggests that SFRP-1 does not inhibit formation and growth of the osteoclast precursors but instead prevents their fusion to form osteoclasts.
The mechanism by which SFRP-1 might inhibit fusion of the mononuclear cells is unknown. However, it might affect specific membrane molecules, including water channel molecules. For example, a recent study indicated that elevated expression of the membrane molecule CD9 was essential for cell fusion during osteoclastogenesis [16], and elevated expression of the water channel molecule aquaporin 9 in osteoclast differentiation was also observed during osteoclastogenesis [17]. In contrast to affecting water channels, SFRP-1 may exert its effect on RANKL. Because SFRP-1 can bind to RANKL [11], this could reduce the functional RANKL concentration and inhibit osteoclastogenesis. Our study showed that the inhibitory effect of SFRP-1 can be reversed by increasing the amount of RANKL in the cultures, implying that SFRP-1 may exert its inhibitory effect through RANKL.
The dental follicle expresses many molecules responsible for osteoclastogenesis and tooth eruption [see review by 1], one of which is osteoprotegerin (OPG), a molecule that inhibits RANKL-dependent osteoclastogenesis. In an earlier study we showed that OPG secreted from dental follicle cells inhibited osteoclast formation in co-cultures of spleen cells and dental follicle cells, and that this inhibitory effect could be reversed by the addition of anti-OPG [18]. The finding in this study that SFRP-1 was expressed in the dental follicle suggested that SFRP-1 may play a role in the inhibition of osteoclastogenesis by the dental follicle. In the in vitro osteoclastogenesis studies, neutralization either by anti-OPG or anti-SFRP-1 partially reduced the inhibitory effect on osteoclastogenesis. However, neutralization by both anti-OPG and anti-SFRP-1 almost completely reversed the inhibitory effect of the follicle cells. This indicated that SFRP-1 contributed to part of the inhibitory effect of the follicle, but was not the sole inhibitor.
The above results further demonstrated that OPG and SFRP-1 may use different pathways to exert their inhibitory effect on osteoclastogenesis. For OPG, it functions as a decoy receptor for RANKL to prevent osteoclastogenesis by inhibiting cell-to-cell signaling [19, 20]. Regarding SFRP-1, its mechanism of inhibition may involve inhibiting fusion of the mononuclear cells, as discussed earlier.
Although our study in rats suggests that SFRP-1 inhibits osteoclastogenesis, just as it appears to do so in mice [11, 15], it may have other functions. In humans, it has been shown that SFRP-1 is expressed in periodontal ligament and gingival cells where it appears to act as an inhibitor of apoptosis [21].
Expression of SFRP-1 in the dental follicle has implications for tooth eruption. As discussed above, the dental follicle expresses two osteoclast inhibitors—OPG and SFRP-1. Our previous studies showed that OPG is down-regulated in the dental follicle of the first mandibular molar at postnatal day 3 by CSF-1 [22, 8]. This study demonstrates that SFRP-1 is down-regulated at both day 3 and day 9. This would favor the major burst of osteoclastogenesis seen at day 3 and the secondary burst of osteoclastogenesis seen at day 10. At day 3 postnatally, RANKL expression is at a normal level in the dental follicle [9], but the levels of OPG and SFRP-1 are low, such that the microenvironment in the dental follicle would favor a major burst of osteoclastogenesis. At day 9, the OPG level is elevated, but the SFRP-1 level is low. In conjunction with a higher RANKL level at this time, the secondary burst of osteoclastogenesis could be promoted. Our future studies will focus on determining what molecule(s) down-regulates SFRP-1 at days 3 and 9 to allow osteoclastogenesis to occur.
Acknowledgments
NIH-R01 Grant DE08911-16 to G.E.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wise GE, Frazier-Bowers S, D’Souza RN. Cellular, molecular, and genetic determinants of tooth eruption. Crit Rev Oral Biol Med. 2002;13:323–34. doi: 10.1177/154411130201300403. [DOI] [PubMed] [Google Scholar]
- 2.Marks SC, Jr, Cahill DR, Wise GE. The cytology of the dental follicle and adjacent alveolar bone during tooth eruption in the dog. Am J Anat. 1983;168:277–89. doi: 10.1002/aja.1001680303. [DOI] [PubMed] [Google Scholar]
- 3.Wise GE, Fan W. Changes in the tartrate-resistant acid phosphatase cell population in dental follicles and bony crypts of rat molars during tooth eruption. J Dent Res. 1989;68:150–56. doi: 10.1177/00220345890680021001. [DOI] [PubMed] [Google Scholar]
- 4.Cielinski MJ, Jolie M, Wise GE, Ando DG, Marks SC., Jr . Colony-stimulating factor-1 (CSF-1) is a potent stimulator of tooth eruption in the rat. In: Davidovitch Z, editor. The biological mechanisms of tooth eruption, resorption and replacement by implants. Birmingham, AL: EBSCO Media; 1994. pp. 429–36. [Google Scholar]
- 5.Wise GE, Lin F, Zhao L. Transcription and translation of CSF-1 in the dental follicle. J Dent Res. 1995;74:1551–57. doi: 10.1177/00220345950740090801. [DOI] [PubMed] [Google Scholar]
- 6.Que BG, Wise GE. Colony-stimulating factor-1 and monocyte chemotactic protein-1 chemotaxis for monocytes in the rat dental follicle. Arch Oral Biol. 1997;42:855–60. doi: 10.1016/s0003-9969(97)00072-1. [DOI] [PubMed] [Google Scholar]
- 7.Steeve KT, Marc P, Sandrine T, Dominique H, Yannick F. IL-6, RANKL, TNF-alpha/IL-1: interrelations in bone resorption pathophysiology. Cytokine Growth Factor Rev. 2004;15:49–60. doi: 10.1016/j.cytogfr.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Wise GE, Yao S, Odgren PR, Pan F. CSF-1 regulation of osteoclastogenesis for tooth eruption. J Dent Res. 2005;84:837–40. doi: 10.1177/154405910508400911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D, Yao S, Pan F, Wise GE. Chronology and regulation of gene expression of RANKL in the rat dental follicle. Eur J Oral Sci. 2005;113:404–09. doi: 10.1111/j.1600-0722.2005.00245.x. [DOI] [PubMed] [Google Scholar]
- 10.Wise GE, Lin F, Fan W. Culture and characterization of dental follicle cells from rat molars. Cell Tissue Res. 1992;267:483–92. doi: 10.1007/BF00319370. [DOI] [PubMed] [Google Scholar]
- 11.Hausler KD, Horwood NJ, Chuman Y, Fisher JL, Ellis J, Martin TJ, et al. Secreted frizzled-related protein-1 inhibits RANKL-dependent osteoclast formation. J Bone Miner Res. 2004;19:1873–81. doi: 10.1359/JBMR.040807. [DOI] [PubMed] [Google Scholar]
- 12.Ramsay G. DNA chips: state-of-the art. Nat Biotechnol. 1998;16:40–44. doi: 10.1038/nbt0198-40. [DOI] [PubMed] [Google Scholar]
- 13.Que BG, Lumpkin SJ, Wise GE. Implications for tooth eruption of the effect of interleukin-1α on nuclear factor-kB gene expression in the rat dental follicle. Arch Oral Biol. 1999;44:961–67. doi: 10.1016/s0003-9969(99)00071-0. [DOI] [PubMed] [Google Scholar]
- 14.Kawano Y, Kypta R. Secreted antagonists of the Wnt signaling pathway. J Cell Sci. 2003;116:2627–34. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 15.Bodine PVN, Zhao W, Kharode YP, Bex FJ, Lambert A-J, Goad MB, et al. The Wnt antagonist secreted frizzled-related protein-1 is a negative regulator of trabecular bone formation in adult mice. Mol Endocrinol. 2004;18:1222–37. doi: 10.1210/me.2003-0498. [DOI] [PubMed] [Google Scholar]
- 16.Ishii M, Iwai K, Koike M, Ohshima S, Kudo-Tanaka E, Ishii T, et al. RANKL-induced expression of tetraspanin CD9 in lipid raft membrane microdomain is essential for cell fusion during osteoclastogenesis. J Bone Miner Res. 2006;21:965–76. doi: 10.1359/jbmr.060308. [DOI] [PubMed] [Google Scholar]
- 17.Aharon R, Bar-Shavit Z. Involvement of aquaporin 9 in osteoclast differentiation. J Biol Chem. 2006;281:19305–09. doi: 10.1074/jbc.M601728200. [DOI] [PubMed] [Google Scholar]
- 18.Wise GE, Yao S, Zhang Q, Ren Y. Inhibition of osteoclastogenesis by the secretion of osteoprotegerin in vitro by rat dental follicle cells and its implications for tooth eruption. Arch Oral Biol. 2002;47:247–54. doi: 10.1016/s0003-9969(01)00109-1. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda H, Shima N, Nakagawa N, Mochizuki SI, Yano K, Fujise N, et al. Identity of osteoclastogenesis inhibitory factor (OCIF) and osteoprotegerin (OPG): a mechanism by which OPG/OCIF inhibits osteoclastogenesis in vitro. Endocrinology. 1998;139:1329–37. doi: 10.1210/endo.139.3.5837. [DOI] [PubMed] [Google Scholar]
- 20.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Goto M, et al. A novel molecular mechanism modulating osteoclast differentiation and function. Bone. 1999;25:109–13. doi: 10.1016/s8756-3282(99)00121-0. [DOI] [PubMed] [Google Scholar]
- 21.Han X, Amar S. Secreted frizzled-related protein 1 (SFRP1) protects fibroblasts from ceramide-induced apoptosis. J Biol Chem. 2004;279:2832–40. doi: 10.1074/jbc.M308102200. [DOI] [PubMed] [Google Scholar]
- 22.Wise GE, Lumpkin SL, Huang H, Zhang Q. Osteoprotegerin and osteoclast differentiation factor in tooth eruption. J Dent Res. 2000;79:1937–42. doi: 10.1177/00220345000790120301. [DOI] [PubMed] [Google Scholar]


