Abstract
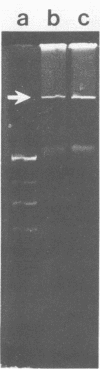
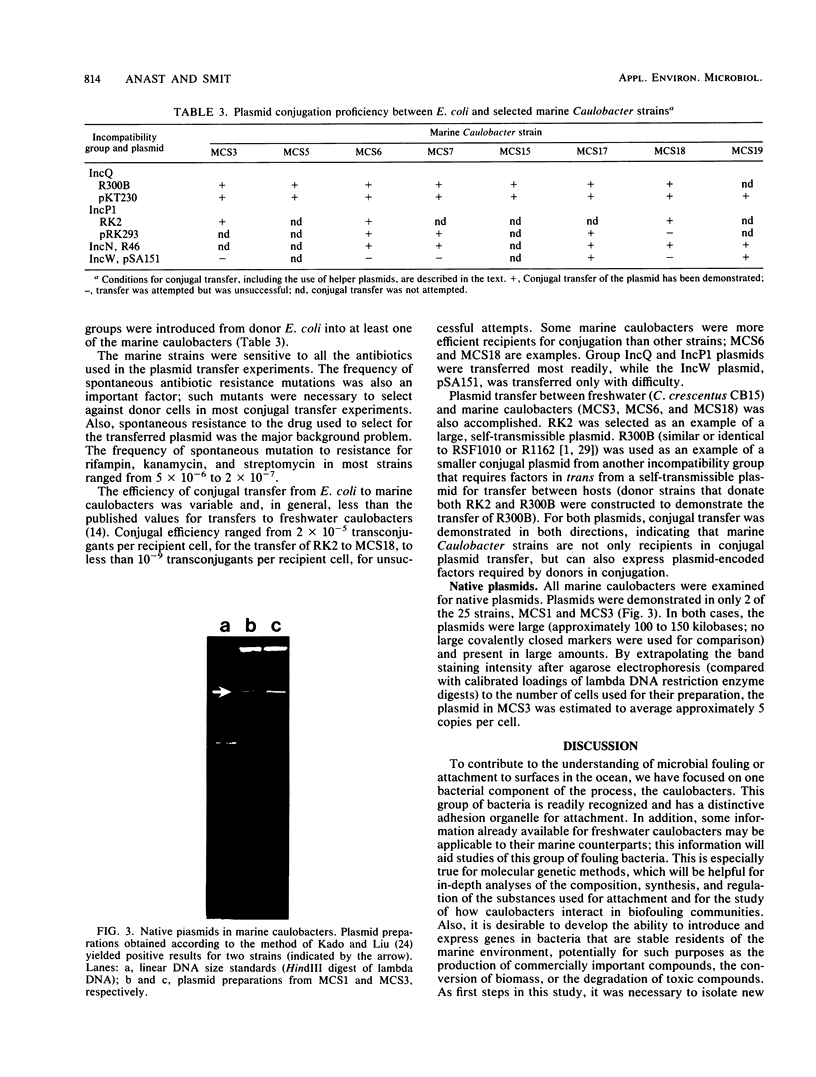
A total of 25 marine caulobacters were isolated from littoral marine sources. Several aspects of their physiology and morphology were examined, as well as their suitability for genetic manipulation in laboratory cultivation. Caulobacters were readily isolated from all sources, including samples from areas containing pollution-related organic compounds. All isolates grew best in media containing seawater, but eight strains grew if sea salts were replaced with NaCl alone, three strains grew at 1/10 the normal sea salt concentration, and one isolate grew, albeit poorly, in freshwater medium. Of the marine isolates, 12 strains grew under anaerobic conditions, indicating that some caulobacters are not obligately aerobic bacteria, as they are currently categorized. Although some freshwater caulobacters are able to oxidize manganese, this capability was not found in these marine caulobacters. Of the marine isolates, 10 strains were resistant to mercury chloride concentrations 10- to 20-fold greater than that tolerated by sensitive bacteria. However, a mercury reductase gene comparable with that found in R100-type plasmids was not detected by gene hybridization. With respect to the potential for genetic experimentation, most strains grew rapidly (3- to 4-h generation time at 30°C), producing colonies on solid media in 2 to 3 days. The isolates were sensitive to antibiotics commonly used in recombinant DNA experiments, and spontaneous drug-resistant mutants were selectable. Conjugal transfer of plasmids from Escherichia coli to several marine caulobacters was demonstrated for four broad-host-range plasmid incompatibility groups, by using both self-transmissible plasmids and cloning-oriented plasmids that require a helper plasmid. Conjugal transfer of broad-host-range plasmids between freshwater and marine caulobacters was also demonstrated in both directions. Native plasmids of approximately 100- to 150-kilobase sizes were found in 2 of the 25 marine Caulobacter strains. The native plasmids were present in relatively high copy number and appeared stable in laboratory culture. In short, the marine caulobacters appeared appropriate as candidates for genetic manipulation and the expression of selected genes in the marine environment.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Barkay T., Fouts D. L., Olson B. H. Preparation of a DNA gene probe for detection of mercury resistance genes in gram-negative bacterial communities. Appl Environ Microbiol. 1985 Mar;49(3):686–692. doi: 10.1128/aem.49.3.686-692.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. T., Croft R. H., Ferber D. M., Gerardot C. J., Schoenlein P. V., Ely B. Genetic mapping with Tn5-derived auxotrophs of Caulobacter crescentus. J Bacteriol. 1982 Aug;151(2):888–898. doi: 10.1128/jb.151.2.888-898.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. T., Rhodes C. S., Ferber D. M., Jenkins B., Kuhl S. A., Ely B. Construction of a genetic map for Caulobacter crescentus. J Bacteriol. 1982 Mar;149(3):889–896. doi: 10.1128/jb.149.3.889-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender R. A. Improved generalized transducing bacteriophage for Caulobacter crescentus. J Bacteriol. 1981 Nov;148(2):734–735. doi: 10.1128/jb.148.2.734-735.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdige D. J., Nealson K. H. Microbial manganese reduction by enrichment cultures from coastal marine sediments. Appl Environ Microbiol. 1985 Aug;50(2):491–497. doi: 10.1128/aem.50.2.491-497.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon F. C., Riedel G. E., Ausubel F. M. Overlapping sequences of Klebsiella pneumoniae nifDNA cloned and characterized. Mol Gen Genet. 1979 Jul 2;174(1):59–66. doi: 10.1007/BF00433306. [DOI] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pJP4. J Bacteriol. 1985 Jan;161(1):466–468. doi: 10.1128/jb.161.1.466-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol. 1982 Feb;149(2):620–625. doi: 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Johnson R. C. Generalized Transduction in CAULOBACTER CRESCENTUS. Genetics. 1977 Nov;87(3):391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Transfer of drug resistance factors to the dimorphic bacterium Caulobacter crescentus. Genetics. 1979 Mar;91(3):371–380. doi: 10.1093/genetics/91.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Vectors for transposon mutagenesis of non-enteric bacteria. Mol Gen Genet. 1985;200(2):302–304. doi: 10.1007/BF00425440. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiorse W. C. Biology of iron- and manganese-depositing bacteria. Annu Rev Microbiol. 1984;38:515–550. doi: 10.1146/annurev.mi.38.100184.002503. [DOI] [PubMed] [Google Scholar]
- Gregory E., Staley J. T. Widespread distribution of ability to oxidize manganese among freshwater bacteria. Appl Environ Microbiol. 1982 Aug;44(2):509–511. doi: 10.1128/aem.44.2.509-511.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Humphreys G. O., Anderson E. S. Molecular studies of R factor compatibility groups. J Bacteriol. 1973 Jul;115(1):387–398. doi: 10.1128/jb.115.1.387-398.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom M. E., Wopat A. E., Nunn D. N., Toukdarian A. E. Manipulation of methanotrophs. Basic Life Sci. 1984;28:319–330. doi: 10.1007/978-1-4684-4715-6_21. [DOI] [PubMed] [Google Scholar]
- Mahler I., Levinson H. S., Wang Y., Halvorson H. O. Cadmium- and mercury-resistant Bacillus strains from a salt marsh and from Boston Harbor. Appl Environ Microbiol. 1986 Dec;52(6):1293–1298. doi: 10.1128/aem.52.6.1293-1298.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R., Figurski D., Helinski D. R. Physical and genetic studies with restriction endonucleases on the broad host-range plasmid RK2. Mol Gen Genet. 1977 Apr 29;152(3):129–135. doi: 10.1007/BF00268809. [DOI] [PubMed] [Google Scholar]
- Meyer R., Hinds M., Brasch M. Properties of R1162, a broad-host-range, high-copy-number plasmid. J Bacteriol. 1982 May;150(2):552–562. doi: 10.1128/jb.150.2.552-562.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E. A., Hynes R. H., Bender R. A. Recombination deficient mutant of Caulobacter crescentus. Mol Gen Genet. 1985;198(2):275–278. doi: 10.1007/BF00383006. [DOI] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter J. S. The caulobacters: ubiquitous unusual bacteria. Microbiol Rev. 1981 Mar;45(1):123–179. doi: 10.1128/mr.45.1.123-179.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J. M., Stanier R. Y. The development of cellular stalks in bacteria. J Cell Biol. 1966 Mar;28(3):423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenlein P. V., Ely B. Plasmids and bacteriocins in Caulobacter species. J Bacteriol. 1983 Feb;153(2):1092–1094. doi: 10.1128/jb.153.2.1092-1094.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L. Generation of polarity during Caulobacter cell differentiation. Annu Rev Cell Biol. 1985;1:173–207. doi: 10.1146/annurev.cb.01.110185.001133. [DOI] [PubMed] [Google Scholar]
- Smit J., Agabian N. Cell surface patterning and morphogenesis: biogenesis of a periodic surface array during Caulobacter development. J Cell Biol. 1982 Oct;95(1):41–49. doi: 10.1083/jcb.95.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Agabian N. Cloning of the major protein of the Caulobacter crescentus periodic surface layer: detection and characterization of the cloned peptide by protein expression assays. J Bacteriol. 1984 Dec;160(3):1137–1145. doi: 10.1128/jb.160.3.1137-1145.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Grano D. A., Glaeser R. M., Agabian N. Periodic surface array in Caulobacter crescentus: fine structure and chemical analysis. J Bacteriol. 1981 Jun;146(3):1135–1150. doi: 10.1128/jb.146.3.1135-1150.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]