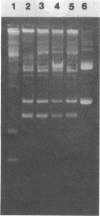
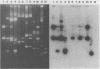
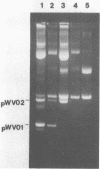
Abstract
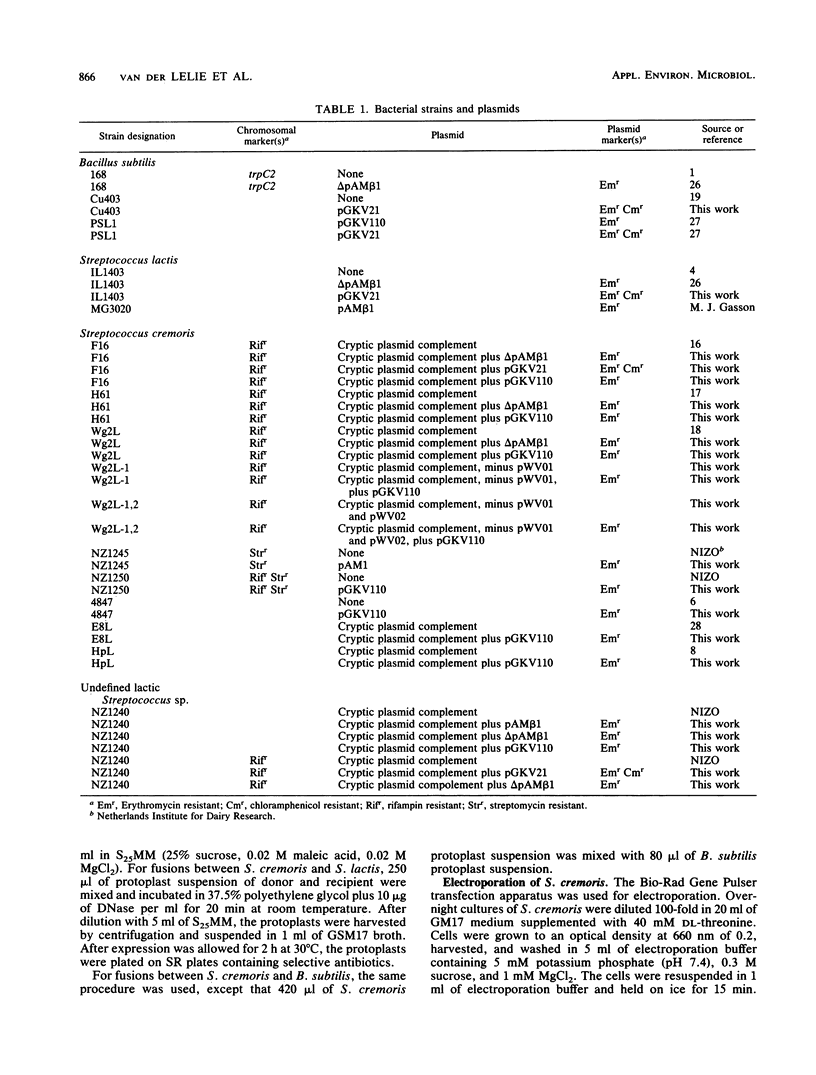
Several Streptococcus cremoris strains were used in protoplast transformation and interspecific protoplast fusion experiments with Streptococcus lactis and Bacillus subtilis, with pGKV110, pGKV21, and ΔpAMβ1 as the marker plasmids. ΔpAMβ1 is a 15.9-kilobase nonconjugative, deletion derivative of pAMβ1, which is considerably larger than the pGKV plasmids (approximately 4.5 kilobases). In general, ΔpAMβ1 was transferred more efficiently than the pGKV plasmids. Using electroporation, we were able to demonstrate that failure of efficient transfer for the pGKV plasmids was, except for one case, caused by incompatibility of these plasmids with resident plasmids of the recipient strain.
Full text
PDF

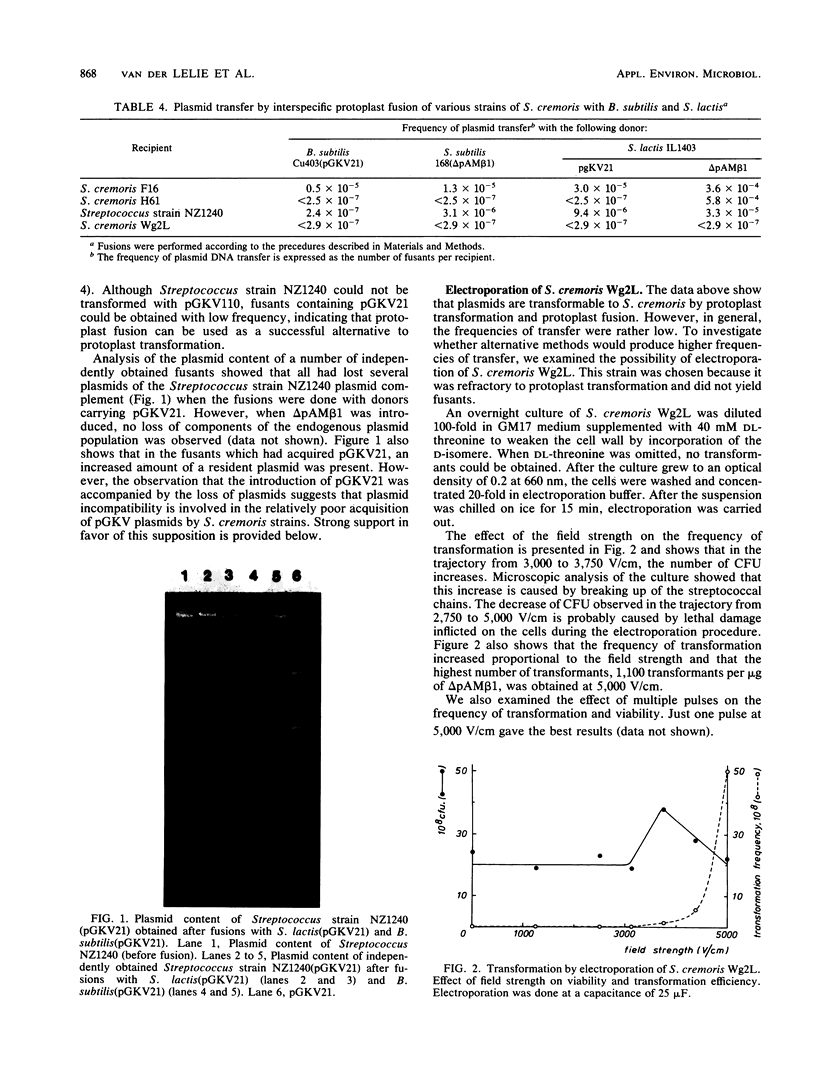

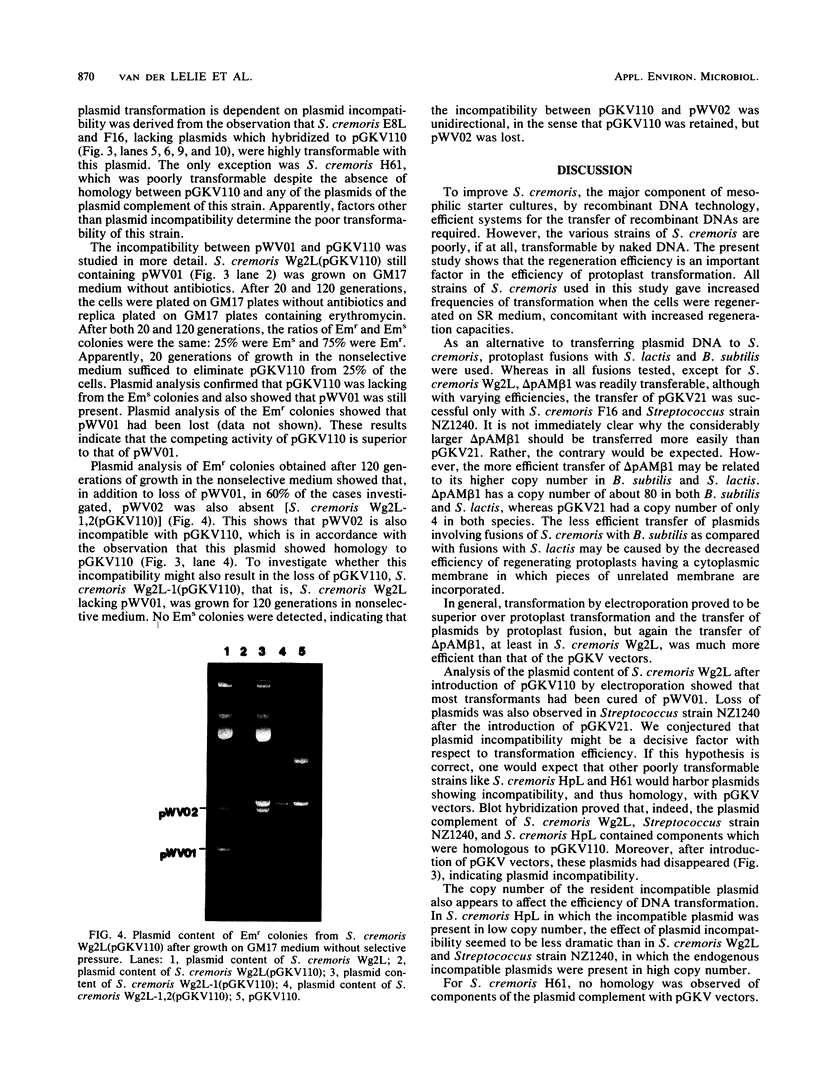

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Qasba P. K. Alkaline transfer of DNA to plastic membrane. Biochem Biophys Res Commun. 1984 Jul 18;122(1):340–344. doi: 10.1016/0006-291x(84)90480-7. [DOI] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo J. K., McKay L. L. Plasmid transformation of Streptococcus lactis protoplasts: optimization and use in molecular cloning. Appl Environ Microbiol. 1984 Aug;48(2):252–259. doi: 10.1128/aem.48.2.252-259.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto R., de Vos W. M., Gavrieli J. Plasmid DNA in Streptococcus cremoris Wg2: Influence of pH on Selection in Chemostats of a Variant Lacking a Protease Plasmid. Appl Environ Microbiol. 1982 Jun;43(6):1272–1277. doi: 10.1128/aem.43.6.1272-1277.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Mendelson N. H., Coyne S. I., Hallock L. L., Cole R. M. Minicells of Bacillus subtilis. J Bacteriol. 1973 May;114(2):860–873. doi: 10.1128/jb.114.2.860-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rottländer E., Trautner T. A. Genetic and transfection studies with B, subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol Gen Genet. 1970;108(1):47–60. doi: 10.1007/BF00343184. [DOI] [PubMed] [Google Scholar]
- Simon D., Rouault A., Chopin M. C. High-efficiency transformation of Streptococcus lactis protoplasts by plasmid DNA. Appl Environ Microbiol. 1986 Aug;52(2):394–395. doi: 10.1128/aem.52.2.394-395.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S., Exterkate F. A., Slangen C. J., de Veer G. J. Comparative Study of Action of Cell Wall Proteinases from Various Strains of Streptococcus cremoris on Bovine alpha(s1)-, beta-, and kappa-Casein. Appl Environ Microbiol. 1986 Nov;52(5):1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelie D., Venema G. Bacillus subtilis generates a major specific deletion in pAM beta 1. Appl Environ Microbiol. 1987 Oct;53(10):2458–2463. doi: 10.1128/aem.53.10.2458-2463.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vossen J. M., Kok J., Venema G. Construction of cloning, promoter-screening, and terminator-screening shuttle vectors for Bacillus subtilis and Streptococcus lactis. Appl Environ Microbiol. 1985 Aug;50(2):540–542. doi: 10.1128/aem.50.2.540-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]