Abstract
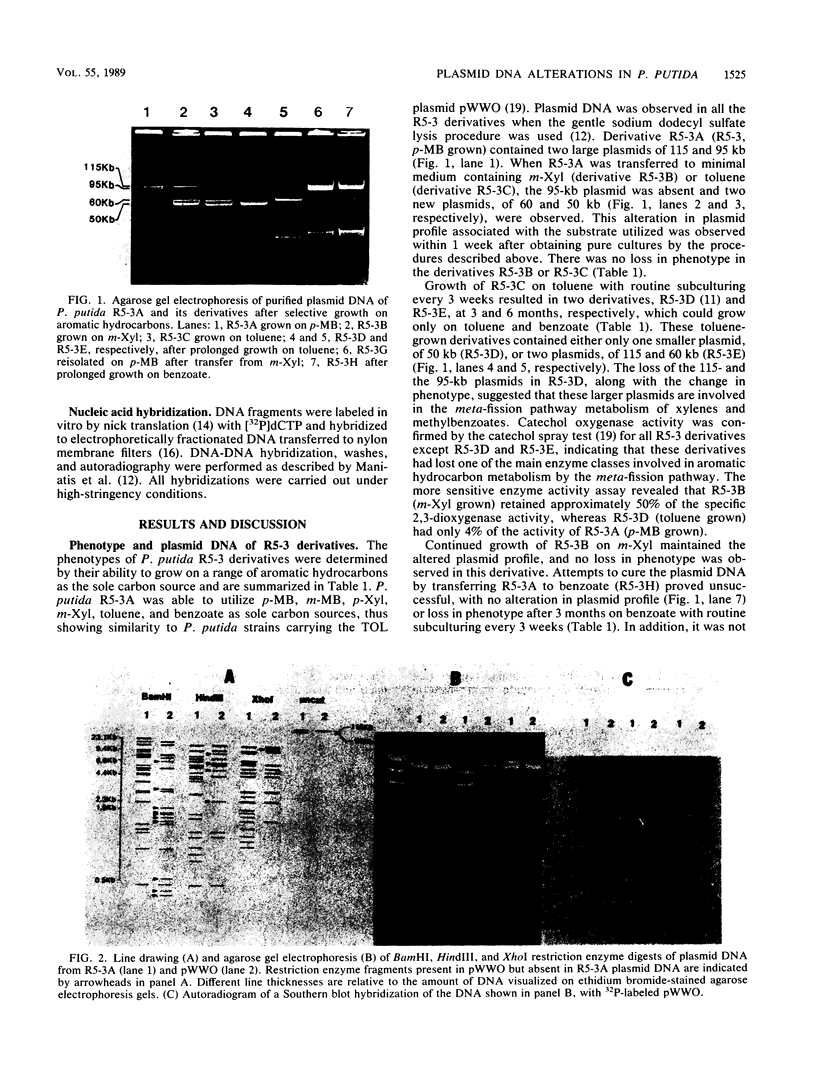
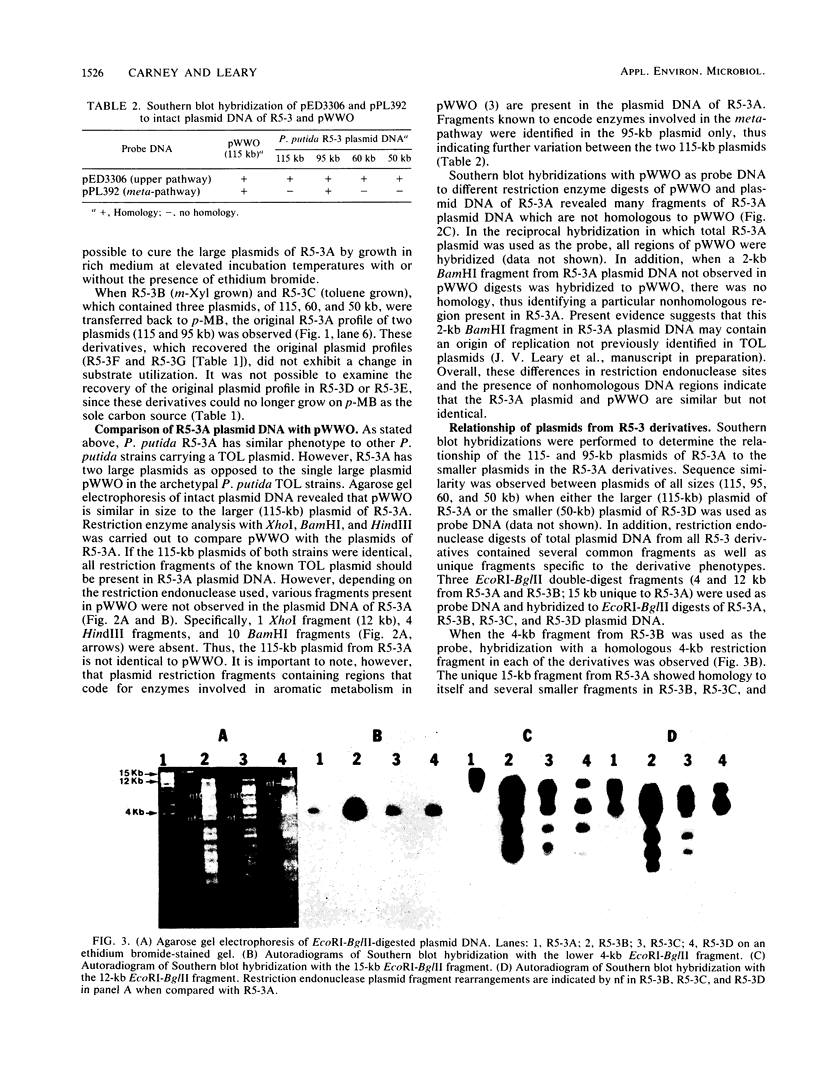
Subcultures of Pseudomonas putida R5-3 altered their plasmid DNA content in specific ways depending on the particular aromatic hydrocarbon utilized as the sole carbon source. Two indigenous plasmids, 115 and 95 kilobases (kb) in size, were observed in R5-3A, which was derived from R5-3 by growth on minimal medium containing p-methylbenzoate as the sole carbon source. When R5-3A was transferred to medium containing m-xylene or toluene, derivative strains were obtained in which the 95-kb plasmid was lost and a new plasmid of 50 or 60 kb appeared. Reversion to the original plasmid profile of R5-3A was observed when xylene- or toluene-grown cells were returned to medium containing p-methylbenzoate. Restriction enzyme analysis and Southern blot hybridizations of total plasmid DNA indicated deletions and rearrangements of DNA restriction fragments in the derivatives maintained on m-xylene and toluene when compared with the original R5-3A. In the derivatives which retrieved the original plasmid profile, the restriction enzyme fragment pattern was identical to that in the original R5-3A, in that the fragments which were missing after growth on m-xylene or toluene were again present. Southern blot hybridizations revealed that part of the plasmid DNA lost from the original plasmid profile was integrated into the chromosomal DNA of xylene-grown R5-3B and that these plasmid fragments were associated with aromatic hydrocarbon metabolism. Hybridization with pathway-specific DNA fragments from the TOL plasmid pWWO indicated that this 95-kb plasmid contains DNA homologous to the meta-fission pathway genes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Lehrbach P. R., Lurz R., Rueckert B., Bagdasarian M., Timmis K. N. Localization and functional analysis of transposon mutations in regulatory genes of the TOL catabolic pathway. J Bacteriol. 1983 May;154(2):676–685. doi: 10.1128/jb.154.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Lehrbach P. R., Timmis K. N. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J Bacteriol. 1984 Oct;160(1):251–255. doi: 10.1128/jb.160.1.251-255.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Leppik R. A., Rekik M., Mermod N., Lehrbach P. R., Reineke W., Timmis K. N. Gene order of the TOL catabolic plasmid upper pathway operon and oxidation of both toluene and benzyl alcohol by the xylA product. J Bacteriol. 1986 Aug;167(2):455–461. doi: 10.1128/jb.167.2.455-461.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth M. E., Shumard D. S., Tatti K. M., Grossman L. I. Rapid purification of yeast mitochondrial DNA in high yield. Biochim Biophys Acta. 1980 Dec 11;610(2):221–228. doi: 10.1016/0005-2787(80)90003-9. [DOI] [PubMed] [Google Scholar]
- Inouye S., Ebina Y., Nakazawa A., Nakazawa T. Nucleotide sequence surrounding transcription initiation site of xylABC operon on TOL plasmid of Pseudomonas putida. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1688–1691. doi: 10.1073/pnas.81.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröckel L., Focht D. D. Construction of chlorobenzene-utilizing recombinants by progenitive manifestation of a rare event. Appl Environ Microbiol. 1987 Oct;53(10):2470–2475. doi: 10.1128/aem.53.10.2470-2475.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sinclair M. I., Maxwell P. C., Lyon B. R., Holloway B. W. Chromosomal location of TOL plasmid DNA in Pseudomonas putida. J Bacteriol. 1986 Dec;168(3):1302–1308. doi: 10.1128/jb.168.3.1302-1308.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staskawicz B. J., Dahlbeck D., Keen N. T. Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6024–6028. doi: 10.1073/pnas.81.19.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valla S., Coucheron D. H., Kjosbakken J. The plasmids of Acetobacter xylinum and their interaction with the host chromosome. Mol Gen Genet. 1987 Jun;208(1-2):76–83. doi: 10.1007/BF00330425. [DOI] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]