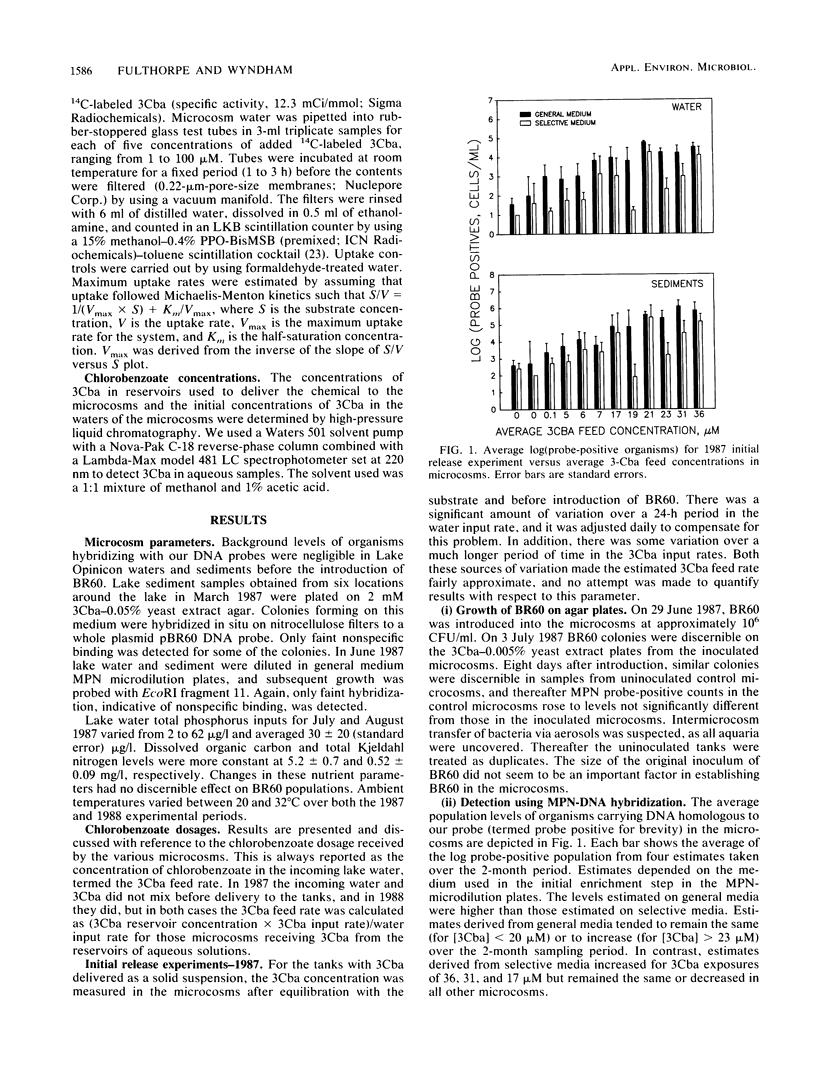
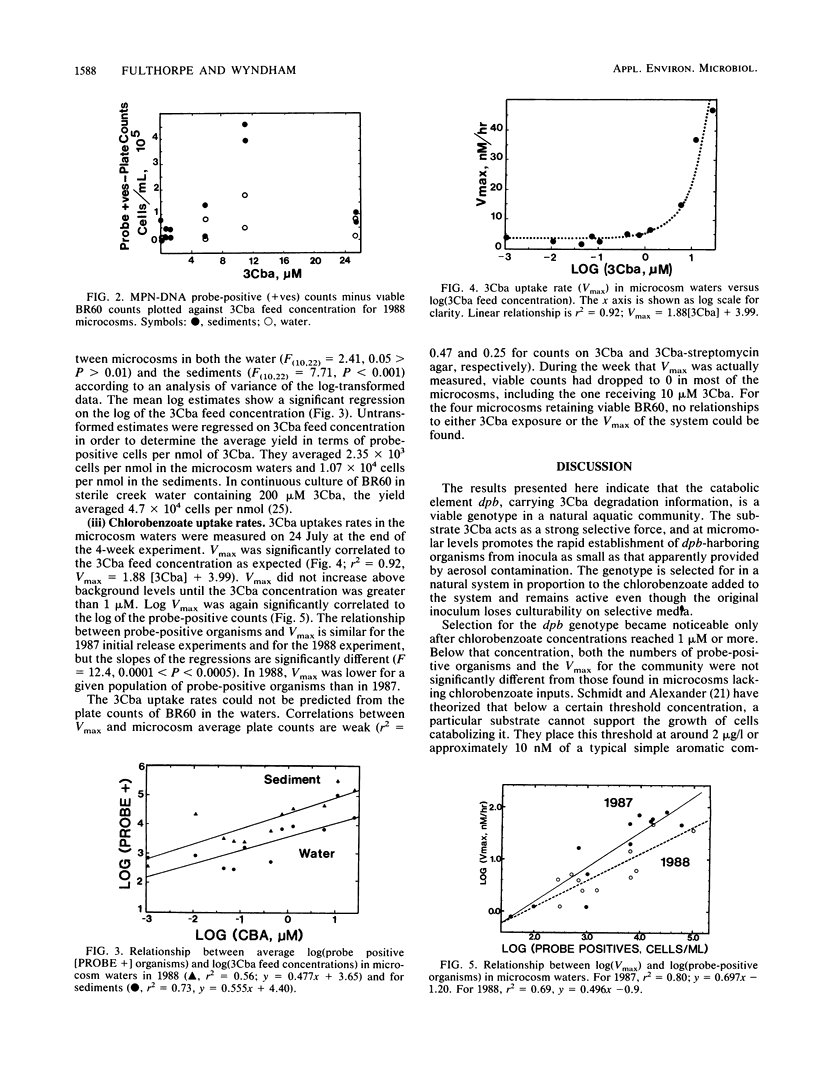
Abstract
A chlorobenzoate-degrading Alcaligenes strain, BR60, was introduced to flowthrough lake microcosms and exposed to 3-chlorobenzoate (3Cba) concentrations from 0 to 25 μM. A DNA probe specific for BR60 chlorobenzoate catabolic genes was used with the most probable number (MPN) technique to enumerate bacteria harboring this genetic information. This MPN-DNA hybridization method combined with [U-14C]3Cba uptake rate measurements allowed the correlation of the size and activity of a specific catabolic population in a natural mixed community for the first time. An experiment involving the release of a streptomycin-resistant strain of BR60 indicated that estimates of bacteria carrying the introduced catabolic genotype often outnumbered plate count estimates of viable BR60 by as much as 3 orders of magnitude, particularly when 3Cba inputs were high. The MPN-DNA hybridization method provided catabolic population estimates highly correlated to 3Cba exposure levels and the [U-14C]3Cba uptake rates in the microcosms. Plate counts of BR60 were poorly correlated with both 3Cba exposure levels and uptake rates. In the absence of chlorobenzoate selection, the catabolic genotype declined to very low levels by the MPN-DNA hybridization technique after 8 weeks in the microcosms.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Edgehill R. U., Finn R. K. Microbial treatment of soil to remove pentachlorophenol. Appl Environ Microbiol. 1983 Mar;45(3):1122–1125. doi: 10.1128/aem.45.3.1122-1125.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Fredrickson J. K., Bezdicek D. F., Brockman F. J., Li S. W. Enumeration of Tn5 mutant bacteria in soil by using a most- probable-number-DNA hybridization procedure and antibiotic resistance. Appl Environ Microbiol. 1988 Feb;54(2):446–453. doi: 10.1128/aem.54.2.446-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R. M., Mallory L. M., Alexander M. Reasons for possible failure of inoculation to enhance biodegradation. Appl Environ Microbiol. 1985 Oct;50(4):977–983. doi: 10.1128/aem.50.4.977-983.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. K., Sayler G. S., Wilson J. T., Houston L., Pacia D. Maintenance and stability of introduced genotypes in groundwater aquifer material. Appl Environ Microbiol. 1987 May;53(5):996–1002. doi: 10.1128/aem.53.5.996-1002.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke D. Use of salicylate to estimate the "threshold" inducer level for de novo synthesis of the phenol-degrading enzymes in Pseudomonas putida strain H. J Basic Microbiol. 1987;27(2):83–89. doi: 10.1002/jobm.3620270206. [DOI] [PubMed] [Google Scholar]
- Karns J. S., Kilbane J. J., Chatterjee D. K., Chakrabarty A. M. Microbial biodegradation of 2,4,5-trichlorophenoxyacetic acid and chlorophenols. Basic Life Sci. 1984;28:3–21. doi: 10.1007/978-1-4684-4715-6_2. [DOI] [PubMed] [Google Scholar]
- Klein T. M., Alexander M. Bacterial inhibitors in lake water. Appl Environ Microbiol. 1986 Jul;52(1):114–118. doi: 10.1128/aem.52.1.114-118.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis D. L., Hodson R. E., Hwang H. M. Kinetics of mixed microbial assemblages enhance removal of highly dilute organic substrates. Appl Environ Microbiol. 1988 Aug;54(8):2054–2057. doi: 10.1128/aem.54.8.2054-2057.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L. N., Sinclair J. L., Mallory L. M., Alexander M. Fate in model ecosystems of microbial species of potential use in genetic engineering. Appl Environ Microbiol. 1982 Sep;44(3):708–714. doi: 10.1128/aem.44.3.708-714.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro P. M., Gauthier M. J., Laumond F. M. Changes in Escherichia coli cells starved in seawater or grown in seawater-wastewater mixtures. Appl Environ Microbiol. 1987 Jul;53(7):1476–1481. doi: 10.1128/aem.53.7.1476-1481.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangnekar V. M. Variation in the ability of Pseudomonas sp. strain B13 cultures to utilize meta-chlorobenzoate is associated with tandem amplification and deamplification of DNA. J Bacteriol. 1988 Apr;170(4):1907–1912. doi: 10.1128/jb.170.4.1907-1912.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- Rubin H. E., Subba-Rao R. V., Alexander M. Rates of mineralization of trace concentrations of aromatic compounds in lake water and sewage samples. Appl Environ Microbiol. 1982 May;43(5):1133–1138. doi: 10.1128/aem.43.5.1133-1138.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerman P. R., Schmidt J. P., Alexander M. Factors affecting the survival and growth of bacteria introduced into lake water. Arch Microbiol. 1988;150(4):320–325. doi: 10.1007/BF00408301. [DOI] [PubMed] [Google Scholar]
- Schmidt S. K., Alexander M. Effects of dissolved organic carbon and second substrates on the biodegradation of organic compounds at low concentrations. Appl Environ Microbiol. 1985 Apr;49(4):822–827. doi: 10.1128/aem.49.4.822-827.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyndham R. C. Evolved aniline catabolism in Acinetobacter calcoaceticus during continuous culture of river water. Appl Environ Microbiol. 1986 Apr;51(4):781–789. doi: 10.1128/aem.51.4.781-789.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyndham R. C., Singh R. K., Straus N. A. Catabolic instability, plasmid gene deletion and recombination in Alcaligenes sp. BR60. Arch Microbiol. 1988;150(3):237–243. doi: 10.1007/BF00407786. [DOI] [PubMed] [Google Scholar]
- Wyndham R. C., Straus N. A. Chlorobenzoate catabolism and interactions between Alcaligenes and Pseudomonas species from Bloody Run Creek. Arch Microbiol. 1988;150(3):230–236. doi: 10.1007/BF00407785. [DOI] [PubMed] [Google Scholar]


