Abstract
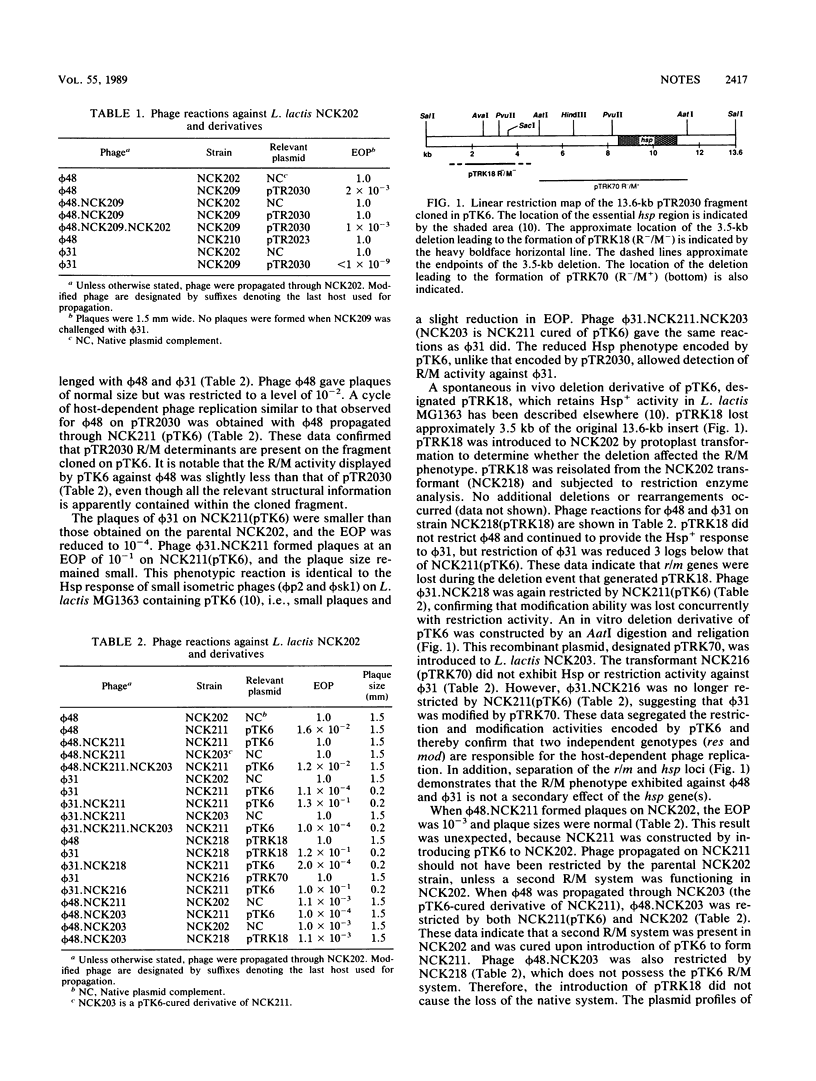
pTR2030 is a conjugative plasmid which encodes resistance to bacteriophage in lactococci by a mechanism that aborts the phage infection (Hsp+). Subcloning and in vivo deletion events showed that two independent mechanisms of resistance are located on a 13.6-kilobase Bg/II fragment cloned in pSA3; one mechanism is responsible for the abortive infection, and the other incodes a restriction modification system. The introduction of pTR2030 or the recombinant plasmid pTK6 resulted in the loss of a resident restriction modification plasmid in Lactococcus lactis NCK202 which was not previously identified.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussemaer J. P., Schrauwen P. P., Sourrouille J. L., Guy P. Multiple modification/restriction systems in lactic streptococci and their significance in defining a phage-typing system. J Dairy Res. 1980 Oct;47(3):401–409. doi: 10.1017/s0022029900021294. [DOI] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Dao M. L., Ferretti J. J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985 Jan;49(1):115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froseth B. R., Harlander S. K., McKay L. L. Plasmid-mediated reduced phage sensitivity in Streptococcus lactis KR5. J Dairy Sci. 1988 Feb;71(2):275–284. doi: 10.3168/jds.S0022-0302(88)79555-7. [DOI] [PubMed] [Google Scholar]
- Gautier M., Chopin M. C. Plasmid-Determined Systems for Restriction and Modification Activity and Abortive Infection in Streptococcus cremoris. Appl Environ Microbiol. 1987 May;53(5):923–927. doi: 10.1128/aem.53.5.923-927.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. L., Sanozky-Dawes R. B., Klaenhammer T. R. Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J Bacteriol. 1988 Aug;170(8):3435–3442. doi: 10.1128/jb.170.8.3435-3442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Romero D. A., McKenney D. S., Finer K. R., Klaenhammer T. R. Localization, cloning, and expression of genetic determinants for bacteriophage resistance (Hsp) from the conjugative plasmid pTR2030. Appl Environ Microbiol. 1989 Jul;55(7):1684–1689. doi: 10.1128/aem.55.7.1684-1689.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Conferred on Lactic Streptococci by the Conjugative Plasmid pTR2030: Effects on Small Isometric-, Large Isometric-, and Prolate-Headed Phages. Appl Environ Microbiol. 1986 Jun;51(6):1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis Audrey W. Conjugal Transfer in Lactic Streptococci of Plasmid-Encoded Insensitivity to Prolate- and Small Isometric-Headed Bacteriophages. Appl Environ Microbiol. 1988 Mar;54(3):777–783. doi: 10.1128/aem.54.3.777-783.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- Murphy M. C., Steele J. L., Daly C., McKay L. L. Concomitant conjugal transfer of reduced-bacteriophage-sensitivity mechanisms with lactose- and sucrose-fermenting ability in lactic streptococci. Appl Environ Microbiol. 1988 Aug;54(8):1951–1956. doi: 10.1128/aem.54.8.1951-1956.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Characterization of Phage-Sensitive Mutants from a Phage-Insensitive Strain of Streptococcus lactis: Evidence for a Plasmid Determinant that Prevents Phage Adsorption. Appl Environ Microbiol. 1983 Nov;46(5):1125–1133. doi: 10.1128/aem.46.5.1125-1133.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Evidence for Plasmid Linkage of Restriction and Modification in Streptococcus cremoris KH. Appl Environ Microbiol. 1981 Dec;42(6):944–950. doi: 10.1128/aem.42.6.944-950.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Phage Resistance in a Phage-Insensitive Strain of Streptococcus lactis: Temperature-Dependent Phage Development and Host-Controlled Phage Replication. Appl Environ Microbiol. 1984 May;47(5):979–985. doi: 10.1128/aem.47.5.979-985.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Leonhard P. J., Sing W. D., Klaenhammer T. R. Conjugal strategy for construction of fast Acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986 Nov;52(5):1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E. Phage resistance in lactic acid bacteria. Biochimie. 1988 Mar;70(3):411–422. doi: 10.1016/0300-9084(88)90215-5. [DOI] [PubMed] [Google Scholar]
- Sijtsma L., Sterkenburg A., Wouters J. T. Properties of the Cell Walls of Lactococcus lactis subsp. cremoris SK110 and SK112 and Their Relation to Bacteriophage Resistance. Appl Environ Microbiol. 1988 Nov;54(11):2808–2811. doi: 10.1128/aem.54.11.2808-2811.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing W. D., Klaenhammer T. R. Conjugal Transfer of Bacteriophage Resistance Determinants on pTR2030 into Streptococcus cremoris Strains. Appl Environ Microbiol. 1986 Jun;51(6):1264–1271. doi: 10.1128/aem.51.6.1264-1271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenson L. R., Klaenhammer T. R. Plasmid heterogeneity in Streptococcus cremoris M12R: effects on proteolytic activity and host-dependent phage replication. J Dairy Sci. 1986 Sep;69(9):2227–2236. doi: 10.3168/jds.S0022-0302(86)80661-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]


