Abstract
Escherichia coli phage P1 Cre recombinase catalyzes the site-specific recombination of DNA containing loxP sites. We report here two crystal structures of a wild-type Cre recombinase–loxP synaptic complex corresponding to two distinct reaction states: an initial pre-cleavage complex, trapped using a phosphorothioate modification at the cleavable scissile bond that prevents the recombination reaction, and a 3′-phosphotyrosine protein–DNA intermediate resulting from the first strand cleavage. In contrast to previously determined Cre complexes, both structures contain a full tetrameric complex in the asymmetric unit, unequivocally showing that the anti-parallel arrangement of the loxP sites is an intrinsic property of the Cre–loxP recombination synapse. The conformation of the spacer is different to the one observed for the symmetrized loxS site: a kink next to the scissile phosphate in the top strand of the pre-cleavage complex leads to unstacking of the TpG step and a widening of the minor groove. This side of the spacer is interacting with a ‘cleavage-competent’ Cre subunit, suggesting that the first cleavage occurs at the ApT step in the top strand. This is further confirmed by the structure of the 3′-phosphotyrosine intermediate, where the DNA is cleaved in the top strands and covalently linked to the ‘cleavage-competent’ subunits. The cleavage is followed by a movement of the C-terminal part containing the attacking Y324 and the helix N interacting with the ‘non-cleaving’ subunit. This rearrangement could be responsible for the interconversion of Cre subunits. Our results also suggest that the Cre-induced kink next to the scissile phosphodiester activates the DNA for cleavage at this position and facilitates strand transfer.
INTRODUCTION
The Cre–loxP recombination system from phage P1 has been the target of extensive biochemical, biophysical and in vivo studies (reviewed in 1). Cre, a member of the integrase family of recombinases, requires no accessory proteins and mediates all steps in site-specific recombination from synapse formation, DNA cleavage, strand exchange, religation, to Holliday junction (HJ) isomerization and resolution (Fig. 1A). It is a very robust enzyme that functions well in a variety of organisms, including bacteria, yeast, plants, insects and mammals. The applied use of Cre allows sophisticated genome engineering, including conditional gene deletions (2), inducible chromosomal translocations (3,4) and other advanced genome manipulations (reviewed in 5,6). Mechan istic studies in a number of laboratories and in particular crystallographic studies by the groups of Van Duyne and Baldwin have shed light on how the enzyme interacts with DNA, as well as the formation of the 3′-phosphotyrosine and the HJ intermediates (7–11). These studies have provided architectural details of these intermediates and have been the basis for a model assuming a subtle, protein-mediated isomerization of the HJ intermediate not involving any branch migration or helical restacking.
Figure 1.
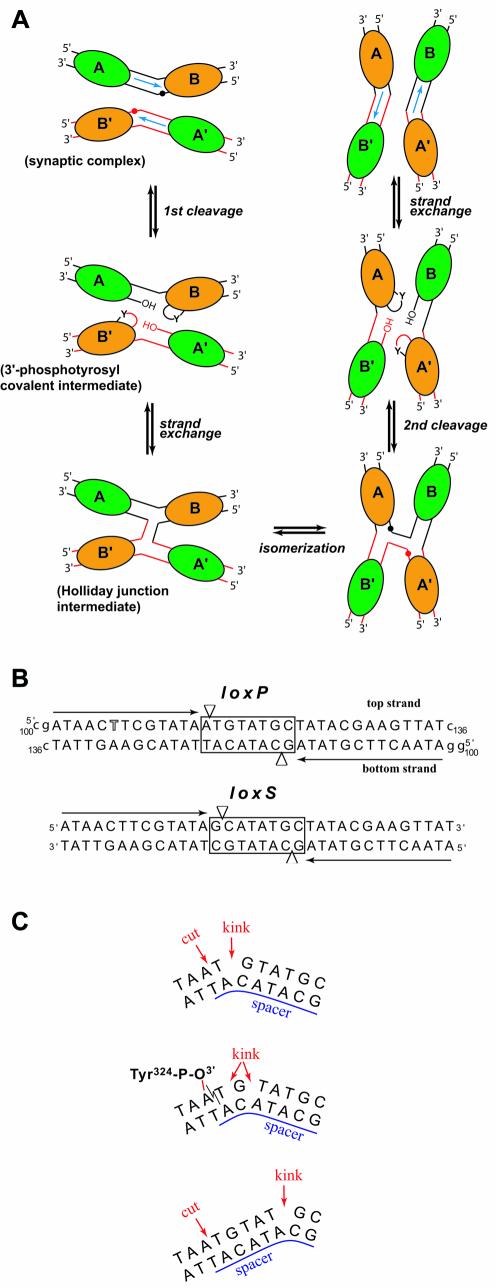
(A) Schematic representation of Cre–loxP-mediated recombin ation. Cre molecules are represented by ellipses (green for non-cleaving subunits and orange for active subunits) and labeled A, B, A′ and B′. Red and black spheres indicate cleavage sites. This figure is adapted from Gopaul et al. (8). (B) Sequences of the loxP (used in this study) and the symmetrized loxS (used by Van Duyne’s group) DNA. The 13 bp inverted repeat sequences are indicated with black arrows. The thymidine position substituted by a 5-iodo-deoxyuridine is highlighted. The spacer region is boxed and arrowheads depict cleavage sites. Residues noted in lower case were added at the 3′ and 5′ ends for crystallization of the complex. (C) In the Cre–loxP synaptic complex, the 8 bp spacer region is kinked immediately next to the top strand cleavage site (top). After the first cleavage, a 3′-phosphotyrosine covalent intermediate is formed (middle). Some rearrangements in the spacer region redistribute the kink over several residues in the cleaved DNA but not all base pairs are still formed. Unlike in the present structures, in the Cre–loxS synaptic complex, the kink is 5 bp away from the cleavage site (bottom).
In the crystal structure of a Cre–loxS synaptic complex containing the inactive R173K or Y324F mutants bound to a symmetrized version of the naturally occurring loxP site (Fig. 1B), an asymmetric bend in the spacer or cross-over region of the DNA was observed involving an atypical large negative roll and positive tilt opening up the major groove (9). This sharp bend is located at the opposite end of the spacer, 5 bp away from the scissile phosphate activated for cleavage (Fig. 1C, bottom). The authors argue, that bending in the left half of the cross-over region stimulates cleavage in the right half and vice versa. Thus, the bending direction would determine, which of the two strands of the spacer will be cleaved first, and thereby provide a structural explanation for the defined order of strand exchange observed for Cre (12–14). However, the crystal structures of Cre–DNA complexes containing modified lox sites with symmetrized spacer regions do not allow direct conclusions concerning the role of the spacer sequence in determining the architecture of the synaptic complex and thereby the outcome of the recombination process. Mutation studies suggest, that the base identity at certain positions of the spacer plays an important role in this process (15).
In a recent study by Baldwin and coworkers, the order of strand exchange was determined from an analysis of the strand composition of the HJ intermediate (11). Their results show that Cre cleaves and exchanges the top strands (also referred to as ‘upper’ strands) first. In the crystal structure of a Cre–loxP HJ complex reported in the same paper, the ‘cleavage-competent’ Cre subunit (also called ‘cleaving’ or ‘activated’ subunit) is bound to the left arm of the junction. Differences in protein contacts to the scissile A and G bases on the left and right hand side of the spacer, respectively, and the orientation of the 198–208 loop region are being proposed as a physical basis for the cleavage order preference by Cre. However, in contrast with this study, a recent re-investigation of the order of strand exchange (16) supports earlier results (12), suggesting that the bottom strand was exchanged first. Clearly, the understanding of such discrepancies needs further investigation.
All crystal structures reported so far of pre-synaptic, synaptic or HJ complexes of Cre are isomorphous, with a crystallographic dyad relating the two Cre-bound DNA duplexes (or opposing strands in the HJ complex) and thereby imposing the 2-fold symmetric, anti-parallel arrangement of the duplexes. We report here two crystal structures of a wild-type Cre–loxP synaptic complex containing the full 200 kDa tetrameric complex in the asymmetric unit, (i.e. without any symmetry imposed by the crystal lattice). One corresponds to a pre-cleavage synaptic complex obtained using a loxP site containing a phosphorothioate modification at the scissile bond that prevents the recombination reaction. The second structure, obtained with an unmodified loxP site is relevant to the synaptic complex after the first cleavage, forming a 3′-phosphotyrosine protein–DNA intermediate. The overall architecture of both complexes is similar to that observed in the loxA and loxS complexes, displaying quasi 2-fold symmetry with an anti-parallel orientation of the two Cre-bound loxP duplexes (see Fig. 3A). Therefore, the pseudo square planar arrangement of the four Cre subunits and DNA arms observed previously is indeed an intrinsic property of the Cre–loxP synapse. A striking feature of the pre-cleavage complex, which clearly distinguishes it from the Cre–loxS structure, is a pronounced kink at the TG/AC step on the left side of the spacer next to the scissile phosphate at the ApT step (Fig. 1C, top). In addition, our 3′-phosphotyrosine intermediate complex, very similar to the Cre–loxA structure, unambiguously shows that the scissile ApT step in the top strand is bound to the Cre subunit displaying an ‘active’ or ‘cleavage-competent’ conformation (Fig. 1C, middle). Our structures therefore suggest that a Cre-induced kink in the wild-type loxP site activates the neighboring scissile phosphate for cleavage. Implications of the Cre-induced spacer conformation and differences between DNA contacts of active and inactive Cre subunits for the mechanism of recombination will be discussed.
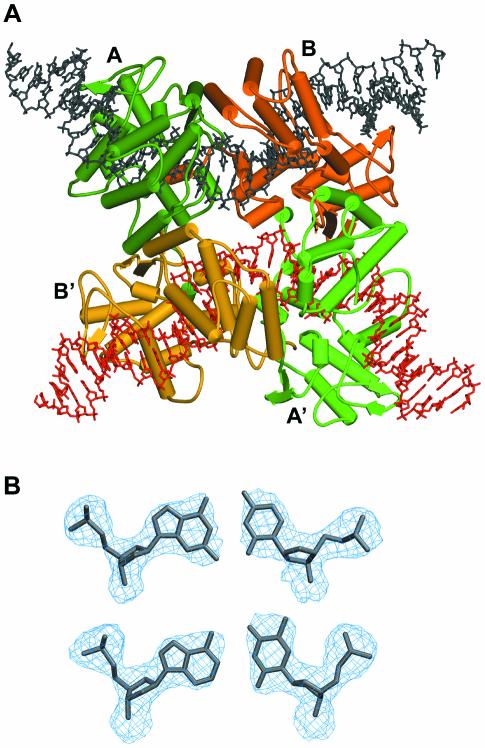
Figure 3.
(A) View of the synaptic tetramer as observed in the asymmetric unit of the present structure. The two DNA duplexes are in black and red. The four Cre proteins subunits are in dark green, light green (non-cleaving subunits), orange and red (cleaving subunits). (B) View of the 3Fobs–2Fcalc electron density map (contoured at 1.4 σ level) around A-T and G-C base pairs showing the overall quality of the electron density map. Note the additional density observed for N2 of guanines compared with adenines and C5 of thymines compared with cytidines.
MATERIALS AND METHODS
Protein and DNA purification
Cre protein was expressed in Escherichia coli from a pGem-derived vector, purified by chromatography on phospho-cellulose P11, Mono S (Pharmacia) and gel filtration (Superdex 200, Pharmacia). As a result of cloning, the expressed protein contained four additional N-terminal residues compared with the SWISSPROT sequence RECR_BPP1: Phe-Gln-Val-Pro. Since the N-terminal tail does not appear in the electron density map, we use the numbering of the published sequence.
DNA oligomers containing the loxP site were chemically synthesized. 37mer oligomers (Fig. 1) without any modifications (oligos 37-5 for ‘top’ strand and 37-9 for ‘bottom’ strand), iodinated (oligo 37-5i, with a subsitution of a deoxy-thymidine into a 5-iodo-deoxyuridine) or containing a non-bridging phosphorothioate modification (mixed R and S diastereomers) introduced either at the ApT (oligo 37-2) or at the GpC (oligo 37-6) steps were used for crystallization experiments. Oligos were dissolved in 100 mM HEPES/NaOH pH 7.6, 50 mM MgCl2, mixed at equimolar ratios and annealed by reducing the temperature from 90 to 25°C in a thermocycler at –0.5°C min–1.
Crystallization and data collection
Crystals were obtained at 20°C with the hanging drop method. Drops (2 µl) contained 0.13 mM Cre, 0.15 mM annealed oligo, 50 mM NaCl, 0.5 mM EDTA, 5% (w/v) glycerol, 50 mM HEPES/NaOH pH 7.6, 75 mM MgCl2 and 12–15% (w/v) PEG 2000 monomethyl ether. For micro-seeding, which strongly increased reproducibility, ∼0.3 µl of a solution containing crystal seeds was added to the drops. Diffraction quality crystals are obtained after 8–12 h only. Crystals were stabilized in a solution containing 20% (w/v) PEG 2000 MME, 100 mM HEPES/NaOH pH 7.6, 100 mM MgCl2 and 20% (w/v) glycerol. Prior to data collection, crystals were flash frozen in liquid ethane. Data were processed with CCP4 (17) and HKL (18) packages.
Structure solution and refinement
The structure was solved by molecular replacement with the program AMoRe (19) using coordinates 1CRX from the protein data bank. A mercury derivative was collected from a crystal soaked for 2 days in a stabilizing solution plus 2 mM ethyl-mercury thiosalicylate (EMTS). Experimental phases up to 4.5 Å resolution were obtained from this dataset with the program SHARP (20) and were used during initial steps of refinement, assuming a 70% solvent content.
Crystallographic refinement was performed with the CNS package (21). Anomalous difference maps were calculated in the 20–3.2 Å resolution range to unambiguously assign the position of both iodine (f” = 2.9 electrons at λ = 0.93 Å) atoms present in the asymmetric unit, revealing two peaks at 8.6 and 7.9 σ above the mean level (these peaks were not present in a structure obtained with a native DNA). As observed previously with a brominated RNA exposed to intense X-ray doses (22), a fast radiolytic cleavage resulting in de-iodination of the uridine occurs during the data collection. In the present case this reaction takes place even more rapidly as the C-I bond is more radiation sensitive than the C-Br bond and due to the extremely long exposure time for data collection (54 s per degree on beamline ID14-2 at the ESRF, Grenoble). As a consequence of this radiolysis, the free iodine does not diffuse in solvent channels, but is rather trapped at the protein–DNA interface, ∼5 Å from the C5 of the uridine (Supplementary Material Fig. S1). Sequence assignment and orientation of the loxP site was further confirmed by using simulated annealing composite omit maps, which show additional density corresponding to N2 of guanines compared with adenines and C5 of thymines compared with cytidines (see Fig. 3B). Several significant peaks appearing in electron density maps were attributed either to water molecules or to magnesium ions depending on the relative charge of the surrounding ligands, their distance, the peak height and the geometry according to (23). To check the identity of cations, attempts at soaking crystal into stabilizing solution containing zinc or manganese were done but systematically lead to crystal cracks.
Coordinates and structure factors for both structures have been deposited in the protein data bank (PDB entries 1nzb and 1ouq).
Recombination assays
For recombination assays, 48mer oligomers were used (5′-CGA TCC GAT AAC TTC GTA TAA TGT ATG CTA TAC GAA GTT ATC TCC GAC-3′ for ‘top’ strand and 5′-GTC GGA GAT AAC TTC GTA TAG CAT ACA TTA TAC GAA GTT ATC GGA TCG-3′ for ‘bottom’ strand). Recombination was performed in the presence of an excess of 37mer oligomer, either without any modification (oligo 37-5/9), with a phosphorothioate on the top strand at the ApT step (oligo 37-2/9), on the bottom strand at the GpC step (oligo 37-5/6), or on both strands (oligo 37-2/6). The reaction buffer contained 20 mM HEPES/NaOH pH 7.6, 2 mM MgCl2, 70 mM NaCl, 5% glycerol, 2 µM of 48mer oligo, 36 µM of 37mer oligo and 37 µM of Cre protein. Reactions were carried out at 39°C for 1 h and stopped by addition of proteinase K digestion (1 mg ml–1 final concentration for 30 min at 37°C or overnight incubation at room temperature) and addition of SDS to 1.25% just before loading samples on the gel. Recombination products were analyzed on a pre-heated denaturing 15% polyacrylamide gel (8 M urea, 45 mM Tris–borate pH 8.3, 2 mM EDTA) and visualized by ethidium bromide staining.
RESULTS AND DISCUSSION
The Cre-induced recombination is blocked by phosphorothioate modification of the loxP cleavage sites
Many enzymes that catalyze phosphodiester bond hydrolysis have a significantly reduced activity on DNA containing phosphorothioate linkages obtained by the substitution of a non-bridging phosphate oxygen by a sulfur (24). It was shown some 15 years ago that the Rp-phosphorothioate modification of the DNA backbone inhibits the cleavage induced by lambda integrase and can be used to determine the order of strand exchange in the recombination process (25,26). As Cre also belongs to the tyrosine recombinase family of site-specific integrases, we decided to use this phosphorothioate strategy to prevent Cre cleavage. As oligonucleotides with a stereo-specific phosphorothioate modification are not available commercially, we used a mixed R and S racemic mixture of a phosphorothioated-modified loxP site, either on the top (at ApT step) or the bottom (GpC step) strand, to check for this hypothesis and to block the first or the second strand Cre-induced cleavage and therefore the recombination process.
Recombination assays performed on loxP sites modified with the introduction of a phosphorothioate at the scissile bonds showed that such DNAs are strongly resistant to protein cleavage (Fig. 2). Despite the use of a racemic mixture of modified DNA, no cleavage was detectable in solution in the timescale of the experiment. At a first sight, one would expect a different impact on the DNA cleavage for S- and R-phosphorothioates, resulting in a partial cleavage inhibition only. Our results show however that both phosphate oxygen atoms seem to be essential for the cleavage, which is in agreement with the active site model of the integrase family proposed by Van Duyne (1). In this model, both oxygen atoms are interacting with essential residues (R173, R292 and W315) and are required for the reaction, as also observed in our structures (see Figs 6 and S3).
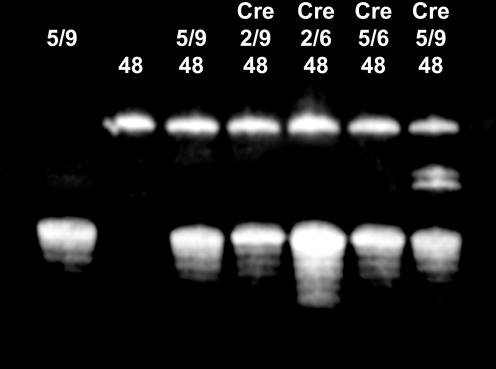
Figure 2.
In vitro recombination experiments performed on native loxP sites or modified by the introduction of a phosphorothioate at the protein cleavage site. Lanes marked ‘48’ contain the 48mer loxP oligomer. Lanes containing the recombinase are indicated with ‘Cre’. The 37mer loxP oligomer is present in lanes marked with ‘5/9’ (unmodified DNA), ‘2/9’ (phosphorothioate on the top strand), ‘5/6’ (phosphorothioate on the bottom strand) and ‘2/6’ (phosphorothioate on top and bottom strands). Due to the presence of overhangs on the 37mer loxP site used for crystallization experiments, two different sizes of recombinant DNA are obtained with unmodified loxP site, corresponding to a 42mer and a 43mer oligomers. No recombinant DNA was detectable in our conditions when a phosphorothioate was present either on the top or the bottom strand.
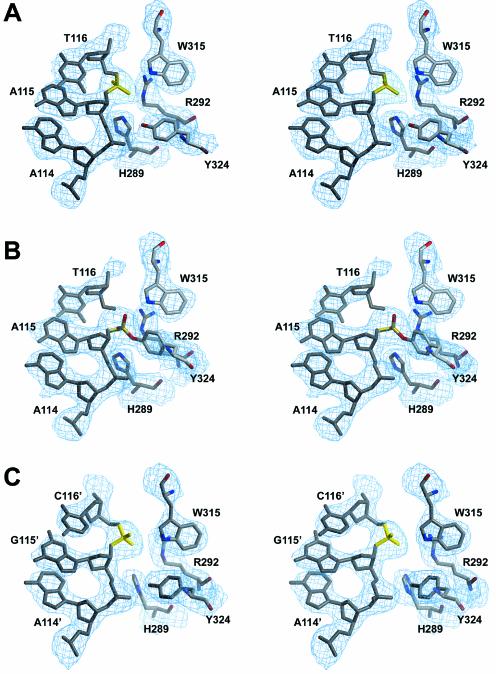
Figure 6.
Stereoview of the active site of Cre recombinase in the Cre–loxP complex. (A) In the cleavage-competent subunit of the pre-cleavage Cre–loxP synaptic complex, (B) the cleavage-competent subunit of the 3′-phosphotyrosine covalent intermediate and (C) in the non-cleaving subunit. The 3Fobs–2Fcalc electron density map contoured at 1.4 σ level is in blue. The scissile phosphate is represented in yellow.
Unexpectedly however, both top and bottom strand modifications seem to have the same effect, blocking any DNA cleavage (Fig. 2). Based on the Cre recombination mechanism (Fig. 1A), one would have rather expected either no cleavage for the modification that prevents the first strand cleavage (the complex remaining a pre-cleavage complex), or 50% of cleavage for the modification blocking the second strand cleavage, as only one strand is exchanged in the HJ intermediate. As discussed below, this result is in agreement with the observation of the same uncleaved Cre–loxP substrate complex in crystals containing either top or bottom strand modification of the DNA. A possible explanation for the lack of observation of any cleavage on the top strand when the bottom strand is modified may be that the first strand cleavage occurs, but quickly reverses to the substrate complex after re-ligation as it cannot proceed to the second strand cleavage due to the phosphorothioate modification. In the same way, we cannot exclude that when the top strand is modified, Cre shortcuts the recombination cycle and cleaves and re-ligates the bottom strand at the GpC step.
The tetrameric Cre synapse is a dimer of dimers with anti-parallel orientation of the loxP duplexes
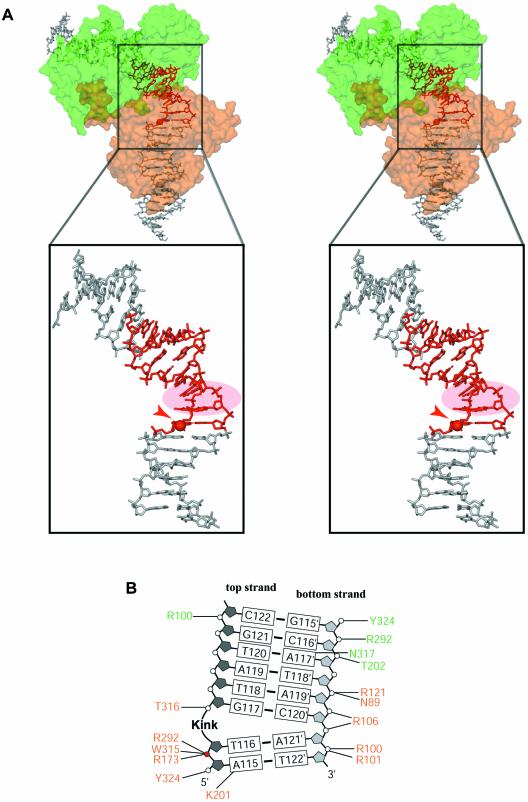
The orthorhombic crystals contain a full ∼200 kDa tetrameric complex in the asymmetric unit, with one Cre subunit bound to each of the inverted repeats of the loxP site duplexes (Fig. 3A). Despite the relatively limited resolution of our structures (2.8 and 2.9 Å resolution, respectively), the anti-parallel orientation of the synapsed loxP sites was clear from the interpretation of electron density maps (Fig. 3B). However, in order to eliminate any ambiguity due to the palindromic sequence with respect to the succession of pyrimidines and purines, we used a modified DNA containing a thymidine to 5′-iodo-deoxyuridine substitution in the inverted repeat region in only one of the strands. Identification of the iodine by anomalous difference maps (see Materials and Methods) confirmed the anti-parallel orientation of the spacer. The quaternary arrangement of the subunits and DNA arms in the synapse is quasi 2-fold symmetric, i.e. subunits A/B and A′/B′ in Figure 3A are related by a local dyad with an overall r.m.s.d. for the Cα-atoms (21–341) of ∼0.4 Å. Distinctly larger deviations (∼2.6 Å) are found for subunits not related by the local dyad (Fig. S2). Differences in the latter pairs are particularly significant in the C-terminal region and the loop regions 198–208 and 314–318 affecting the positions of lysine 201, a residue reported to be catalytically important (9), the active site tyrosine 324 and tryptophane 315. In addition, there is some change in the relative orientation of the N- and C-terminal domains corresponding to a rotation of ∼10°.
The synapsed loxP sites are oriented anti-parallel with the four Cre-bound arms forming a HJ-like, pseudo square- planar array. A similar overall architecture was found in the Cre–loxA and loxS as well as the Cre–loxP HJ complexes. However, in the latter structures, an exact 2-fold symmetry is imposed by the crystal lattice. Our structure containing a wild-type loxP site clearly shows, that the molecular 2-fold symmetry with an anti-parallel orientation of the DNA duplexes is an intrinsic property of the Cre–loxP recombination synapse.
Identification of two different Cre–loxP structures: a pre-cleavage complex and a 3′-phosphotyrosine covalent intermediate
An uncleaved complex could unambiguously be identified from several data sets obtained with a DNA containing a phosphorothioate modification either at top or bottom strand scissile bonds. Indeed, although they interfere with different cleavage steps, both modifications lead to the same crystal structure. As our recombination experiments clearly showed that the introduction of a phosphorothioate at scissile bonds prevents recombination, and given the short timescale required for crystallization (see Materials and Methods), we assumed that these structures correspond to the pre-cleavage synaptic complex, and not to a recombination product. Since the best diffracting crystal was obtained for a modification on the bottom strand of the DNA, such a crystal was used for refinement in Table 1. It shows an extremely well-defined electron density map for the protein as well as for the DNA, including the complete spacer region (Fig. 4A). However, due to the relatively limited resolution and the use of a racemic mixture of R- and S-phosphorothioates, it was not possible to observe the sulfur either on Fobs–Fcalc difference density maps or on anomalous difference maps.
Table 1. Data collection and refinement summary.
| Synaptic complexa,b |
Covalent intermediatea |
|
| Space group | P212121 | P212121 |
| Unit cell (Å) | A = 107.8, b = 161.1, c = 195.7 | a = 108.9, b = 164.2, c = 194.7 |
| X-ray source | ESRF ID29 | SLS PX |
| Max. resolution (Å) | 2.8 | 2.9 |
| Completenessc (%) | 97.4 (94.6) | 96.2 (98.2) |
| Rsymc (%) | 7.3 (31.8) | 6.7 (33.0) |
| R factor (%) | 23.0 | 23.0 |
| Rfree factor (%) | 25.2 | 26.2 |
| r.m.s.d. bonds length (Å) | 0.007 | 0.020 |
| r.m.s.d. bonds angles | 1.2° | 1.6° |
| Estimated coordinate error (Å) | 0.41 | 0.45 |
aStructure obtained with an iodinated loxP DNA.
bPhosphorothioate modification of the loxP DNA at the cleavage site on the bottom strand.
cValues in parenthesis correspond to percentage in outermost shell.
Figure 4.
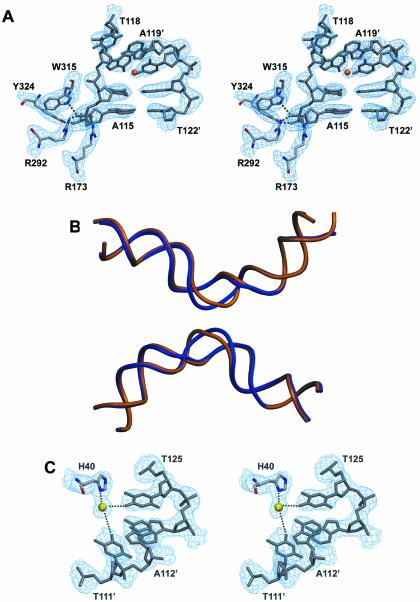
(A) Stereoview of the 3Fobs–2Fcalc electron density map (contoured at 1.4 σ level) around the active site of the ‘cleavage-competent’ Cre subunit and the kink region of the loxP site. A metal ion (possibly a magnesium) is indicated by an orange sphere. (B) DNA backbone superposition of the wild-type loxP sequence as observed in the present structure (in blue) and the symmetrized loxS sequence (PDBID 4CRX; the second duplex was generated by applying 2-fold symmetry). While the overall bending is the same, marked differences are found in the spacer region resulting from different locations of kinks. (C) Stereoview of the 3Fobs–2Fcalc electron density map (contoured at 1.4 σ level) showing a magnesium-mediated protein–DNA contact.
The 3′-phosphotyrosine covalent intermediate was identified during the refinement of several data sets obtained from crystals containing unmodified DNA. These crystals display a significant lack of isomorphism compared with previous data sets (see Table 1). Fourier difference density maps, combined with simulated annealing composite omit maps revealed a major movement (∼3 Å) of the C-terminal part of the activated Cre subunit, which contains the attacking tyrosine. In particular, an extremely short distance between the Tyr324 hydroxyl group and the cleavable phosphate at the ApT step of the top strands was observed (<2 Å), correlated with a strong negative density peak in the difference density map for the sugar-phosphate backbone at this position. In addition, His289, a residue belonging to the active site of the protein that could act as a proton donor for the Tyr324 leaving group (7), is directly contacting the scissile phosphate as observed previously in the Cre–loxA covalent intermediate, but not in Cre–loxP, Cre–loxS or Cre–HJ intermediate complexes (see below). Finally, a superposition of this structure with all available Cre–DNA complexes shows that this structure only fits with the Cre–loxA suicide substrate (see below) also containing a 3′-phosphotyrosine covalent intermediate (7). As the isomerization of the complex is not possible given the DNA-driven crystal packing, and as the activated Cre subunit is oriented in the same way than in the pre-cleavage complex, we assumed that this structure is relevant for the first cleavage step, and not the second one. Noticeably, no uncleaved complex was detectable on electron density maps, even using low contour levels. We were however not able to detect this covalent intermediate by gel electrophoretic mobility assays using dissolved crystals, probably due to the reversibility of the reaction in solution.
Interestingly, using the same wild-type Cre and a similar loxP DNA substrate, Baldwin and coworkers obtained a Cre–HJ intermediate structure (11). This different result might be attributed to the presence in our crystallization conditions of divalent cations (75 mM MgCl2) that promote the recombination process, but are not essential (27). Another important difference in crystallization conditions concerns the pH used: 5.0–5.5 in the Cre–HJ intermediate instead of 7.6 in the present structure. This difference in pH can affect the protonation of some important amino acids, and in particular of the His289 located in the active site. In agreement with this hypothesis, the crystallization conditions used for the Cre–HJ intermediate prevent cleavage and a 10-min pre-incubation was necessary for the reaction to take place (11).
DNA bending and conformation of the spacer in the Cre–loxP synapse: comparisons with Cre–loxA and Cre–loxS complexes
A global superposition with the Cre–loxA and Cre–loxS complexes highlights the overall similarity of the quaternary arrangement (Fig. 4B). Superimposing the A and B subunits of the Cre–loxP structure with the corresponding subunits in the loxA and loxS complexes using the Cα-atoms of residues 20–323 results in an r.m.s.d. of 1.9 and 1.8 Å, respectively. The interfaces between subunits are also mostly conserved. Likewise, the specific interaction of the Cre subunits with the 13 bp inverted repeats is essentially the same in these structures, with the N-terminal domain contacting the DNA in the major groove on one side, the C-terminal domain in adjacent major and minor grooves on the opposite side. But in contrast to previously reported Cre complexes, several metal ion-mediated protein–DNA contacts are observed in these regions (Fig. 4C), most likely due to the presence of large amounts of magnesium in our crystallization conditions. Like in the loxA and loxS complexes, direct protein contacts to the bases in the spacer region are rare. A common feature is also the distinctly different protein–phosphate backbone contact pattern for the top and bottom strands (see Discussion below).
The overall bending of the DNA duplexes in the Cre–loxP synapse is rather similar to the one in Cre–loxA and Cre–loxS complexes, with an angle of ∼100° between the left and right hand arms (Fig. 4B). However, this bend is achieved by a distinctly different conformation of the spacer regions. In the wild-type Cre–loxP pre-cleavage complex, a pronounced kink at the TG/CA step next to the scissile phosphate (Figs 4A and 5) contributes roughly half to the overall bending. A positive roll of ∼45° leads to an unstacking of bases at this step and a massive widening of the minor groove (Figs 4A and 5A). In contrast, bending in the symmetrized loxS synapse is primarily due to a kink 5 bp away at the other end of the spacer (Fig. 1C, bottom). The kink observed in the loxS complex, which is associated with a large negative roll and positive tilt, an opening of the major and compression of the minor groove, is atypical and not normally found in protein–DNA complexes. The striking differences between the symmetric loxS and the wild-type loxP spacer conformations have at least in part to be attributed to the different nucleotide sequences within the spacer regions. As a consequence of the different spacer conformations we do not see the unfavorable close approach of two phosphates in the center of the spacer observed in the Cre–loxS complex. It was speculated by the authors that the energy associated with a stereochemically strained spacer conformation promotes the subsequent strand exchange step (9). On the basis of the loxP complex, we would instead argue, that a Cre-induced kink activates the neighboring scissile phosphate for cleavage and facilitates dissociation of the cleaved strand during strand exchange. In this context it is interesting to note that divalent cations, although not essential, do promote the Cre-catalyzed recombination considerably (27). Interestingly, it was proposed that solvated divalent cations promote and stabilize unstacked conformations by interactions with the nucleobase π electrons (28). Stabilization of a kinked DNA structure together with an overall electrostatic shielding effect, may therefore explain the observed acceleration of Cre-catalyzed recombination in the presence of divalent cations.
Figure 5.
(A) Stereoview of a Cre dimer (represented by the solvent accessible surface of the protein) interacting with a loxP site (the spacer region is in red). The ‘cleavage-competent’ Cre (in orange) is bound close to the top strand cleavage site (ApT step). A close-up view of the spacer region shows that a sharp kink (circled in pink) is located at the TG/CA step next to the scissile phosphate in the top strand. The scissile bond is represented by a red sphere and indicated by a red arrowhead. (B) Schematic drawing of protein–DNA contacts in the spacer region. Amino acids are colored in green or orange depending on whether they belong to the ‘non-cleaving’ or the ‘cleavage-competent’ subunit, respectively. Note that except for the scissile phosphate, very few contacts are formed with the top strand, facilitating its exchange after cleavage. Nucleotide numbering follows the one used in PDB entries. Top and bottom strands are depicted in dark and light gray, respectively. The scissile phosphate of the top strand is indicated as a red sphere.
The spacer conformation in the 3′-phosphotyrosine Cre–loxP covalent intermediate is somewhat different to the one observed in the pre-synaptic complex. Indeed, the 4 nt (5′G117TAT1203′) following the residue next to the cleavage point are poorly defined in electron density maps, very likely due to the presence of two conformations of this part of the spacer region. It is however clear from omitted-refined electron density maps that T116 following the cleavage point remains base-paired and that the kink is more distributed than in the pre-cleavage complex, in particular between the T116/G117 and G117/T118 steps (Fig. 1C, middle). Surprisingly, nucleotides belonging to the opposite strands (5′A117′ TAC120′3′) are well defined in electron density maps, which helped to analyze the spacer conformation. Noticeably, alternate conformations of the same nucleotides were also observed for the Cre–loxP HJ complex (11). This spacer conformation is therefore different to the one observed in the Cre–loxA covalent complex, where the cleaved strand lacking the 5′-cytidine (which diffuses into the solvent following the cleavage) was completely unpaired in the spacer region and moved towards the middle of the synaptic complex for initiating strand exchange (7). On the contrary, our 3′-phosphotyrosine covalent intermediate containing the wild-type loxP site shows a subtle re-arrangement of the spacer region associated with a redistribution of the kink, rather than a melting of the cleaved strands. As the melting is necessary for the strand exchange, it could therefore be relevant for the earliest step of the strand exchange.
Differences in protein contacts of top and bottom strands in the spacer region
The Cre-induced asymmetric bending and kinking in the spacer region of the loxP recombination site and the concomitant conformational changes in the protein subunits lead to differences in the protein contact pattern of the top and bottom strands of the spacer. The only direct base contacts in the spacer involve the scissile adenine and guanine bases at the 5′ ends of the spacer in the top and bottom strands. While both bases are contacted by K86 from the major groove side (via N7 in case of adenine; via O6 and N7 in case of guanine), only the scissile adenine in the top strand is contacted by the catalytically important K201 from the minor groove via N3 (Fig. 5B). This base-specific contact is well defined in electron density maps of both the Cre–loxP pre-cleavage and 3′-phosphotyrosine covalent intermediate complexes. Interest ingly, this contact is also present in the Cre–loxP HJ intermediate, but not in Cre–loxA and Cre–loxS synapses, where this lysine was disordered (1). Furthermore, no such contact is present for the ‘non-cleaving’ subunits due to differences in the conformation of loop 198–208. Therefore, as already suggested for the Cre–loxP HJ complex (11), this residue might be responsible for the cleavage preference of Cre and the specific recognition of the adenine present in the left arm of loxP through this base-specific contact.
With the exception of active site residues, there are almost no contacts to the backbone of top strands in the spacer region, facilitating their exchange after cleavage (Fig. 5B). In contrast, bottom strands are more tightly bound to the protein, e.g. via a strong salt bridge between R121 and the central phosphate of the spacer (P119′), as well as ionic contacts of arginines 100, 101 and 106 to the neighboring phosphates P120′, P121′ and P122′, respectively (Fig. 5B).
Implications for the mechanism and order of strand exchange in Cre–loxP site-specific recombination
Like the previously published Cre complexes, structures of the synaptic wild-type Cre–loxP pre-cleavage and 3′-phosphotyrosine intermediate complexes reported here strongly support a cis-cleavage mechanism, where residues of the catalytically essential RHR triad (R173/H289/R292), as well as the attacking tyrosine nucleophile (Y324), belong to the same subunit. In the pre-cleavage complex, dispositions of the active site residues are similar in the ‘non-cleaving’ and ‘cleavage-competent’ Cre subunits. There are however some distinct differences to be noted. In the ‘non-cleaving’ subunits the Y324-OH nucleophile is 5 Å away from the scissile phosphate and directly interacts with the adjacent phosphate (Figs 6A and S3A). In the ‘cleavage-competent’ subunits, the distance between the scissile phosphate and the tyrosine is significantly reduced to 4 Å, while the contact to the adjacent phosphate is maintained (Figs 6C and S3C). Another difference is found for H289, which possibly acts as a general base for accepting the Y324 hydroxyl proton (1). The distance between the Nε position of H289 and the scissile phosphate is 4.7 Å in ‘non-cleaving’ subunits, but reduced to 4.0 Å in ‘cleavage-competent’ subunits. The movement of the catalytically important K201 as a consequence of rearrangements in the loop 198–208, as well as the changes in the relative orientations of the N- and C-terminal domains, have already been discussed.
In the Cre–loxP 3′-phosphotyrosine covalent intermediate, the active site of the cleaving subunits undergoes several significant changes identical to the one found in the Cre–loxA covalent intermediate (7). As expected, the most important change concerns the attacking Y324, which moves towards the scissile bond and is covalently linked to the DNA (Figs 6B and S3B). Related to this movement, a major conformational change is observed for the helix containing Y324 (helix M according to 7), which moves 3 Å towards the DNA (Fig. 7A). This motion is propagated towards the C-terminal part of the protein, including helix N, which binds in a hydrophobic pocket of the neighboring Cre monomer. In addition, the reorientation and the motion of the scissile phosphate following the cleavage allows a direct interaction (2.9 Å) between the latter and the Nε position of H289. This rearrangement of the scissile phosphate also permits the two phosphate oxygens to be correctly oriented for interacting with H289 and W315 (Figs 5B and S3B).
Figure 7.
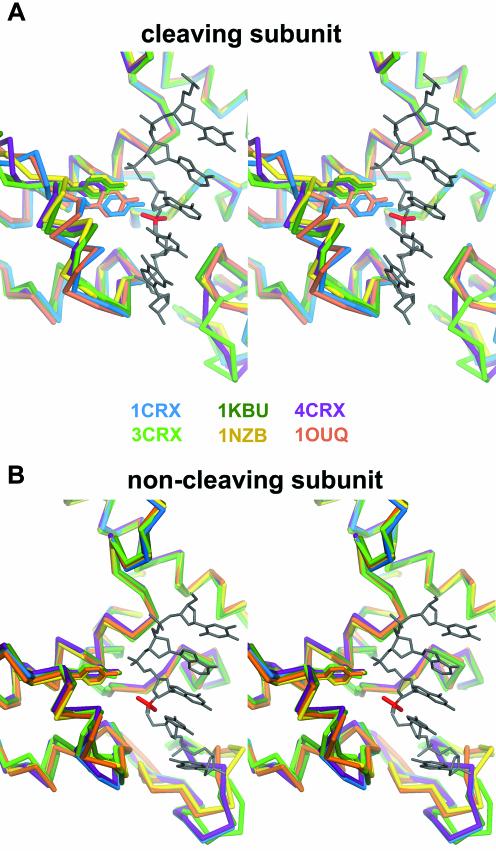
Stereoviews showing a superposition of the active site of several Cre–DNA complexes for the ‘cleavage-competent’ (top) and ‘non-cleaving’ (bottom) subunits. Proteins are colored as follows: purple, Cre–loxS synaptic complex (4CRX); light green, Cre-immobile HJ intermediate complex (2CRX); dark green, Cre-wild-type HJ intermediate complex (1KBU); blue, Cre–loxA suicide substrate (3′-phosphotyrosine covalent intermediate); yellow, Cre–LoxP synaptic complex (this study, 1nzb); orange, Cre–loxP 3′-phosphotyrosine covalent intermediate (this study, 1ouq).
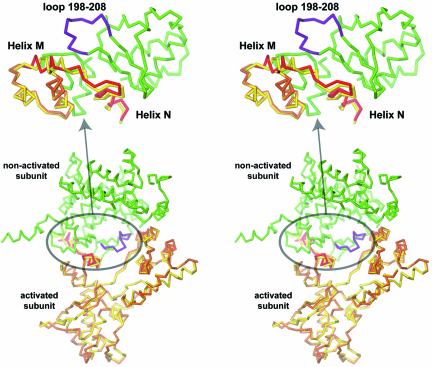
Unlike ‘cleavage-competent’ subunits, the ‘non-cleaving’ subunits in the 3′-phosphotyrosine covalent intermediate and the pre-cleavage state complexes are essentially identical (Fig. 7B). But noticeably, the moving region of the ‘cleavage-competent’ subunits is in the neighborhood of the loop 198–208 of the ‘non-cleaving’ subunits and might therefore be involved in the communication between the two subunits and the conformational switch interconverting cleaving- and non-cleaving Cre subunits (Fig. 8).
Figure 8.
Stereoviews of a Cre dimer showing inter-subunits contacts in the neighborhood of the active site. The non-cleaving subunit is in green and its loop 198–208 in purple. Cleavage-competent subunits before cleavage (in yellow) and in the 3′-phosphotyrosine covalent intermediate (in orange) are superimposed. The moving C-terminal part containing helices M and N is in red. The close-up view (top) is from a different point of view to highlight the movement of helices M and N.
Noticeably, a superposition of ‘cleavage-competent’ subunits of all solved crystal structures shows that they are nearly identical for loxP, loxS synaptic complexes as well as for the two HJ complexes, whereas the loxA and loxP covalent intermediates both display the same movement in their C-terminal part containing helices M and N (Fig. 7A). This is not the case for ‘non-cleaving’ subunits, which are isomorphous in all Cre complexes (Fig. 7B).
What does the Cre–loxP complex tell us about the order of strand cleavage and exchange? While some members of the Int family of recombinases, like e.g. Flp, do not seem to have a defined order of strand cleavage, Cre has an intrinsic bias for initiating the recombination preferentially on one strand. So far, two alternative models have been proposed, depending on whether the top or the bottom strand is cleaved and exchanged first. Two independent investigations (12,16) suggest that the bottom strand is cleaved first at the GpC step. However, the alternative order of strand exchange was observed by an analysis of strand composition of HJ intermediates (11), suggesting that the top strand is cleaved first, i.e. the ApT step at the left hand side of the loxP site. In line with the latter cleavage bias, we find in the pre-cleavage synaptic complex the ‘cleavage-competent’ subunits B/B’ associated with the ApT step in the top strand on the left hand side of the spacer. Moreover, the observation of the 3′-phosphotyrosine covalent intermediate shows that the conformation of the Cre–loxP complex captured in our crystals is catalytically relevant and not a crystallization artefact due to a wrong spacer conformation. However, our crystallographic results are clearly not compatible with an initiation of the recombination on the bottom strand and we do not have any explanation about the discrepancy with results reporting this cleavage bias.
By introducing bulges consisting of three additional adenine nucleotides at specific positions in one strand of the spacer and thereby forcing the DNA to bend in a given direction it was shown that the covalent intermediate tended to accumulate on the bulge strand (14). Furthermore, these experiments demonstrated that a bulge next to or 1 bp away from the cleavage site is most effective in determining or changing the cleavage bias. These experiments are in excellent agreement with our crystallographic results. First, they provide clear evidence, that the DNA bend direction determines the order of cleavage events. Secondly, the position-dependence of the bulge-effect strongly supports our conclusion that the observed kink neighboring the scissile bond activates for cleavage.
CONCLUSIONS
The crystal structure of a wild-type Cre–loxP synapse with a tetrameric complex in the asymmetric unit shows that the near 2-fold symmetric, anti-parallel arrangement of the loxP sites is an intrinsic property of the Cre recombination synapse. The overall architecture and bending of the DNA is rather similar to that of the previously determined loxA, loxS and HJ complexes displaying the same pseudo square-planar arrangement of the Cre subunits and DNA arms. A pre-cleavage complex was trapped using a loxP site modified by the introduction of a phophorothioate at scissile bonds, a modification that prevents the recombination reaction. It shows that the ‘cleavage-competent’ Cre subunit is bound to the left hand side of the loxP spacer region. A pronounced kink of the DNA is found at the neighboring TG/AC step associated with a large positive roll and a widening of the minor groove. Using a non-modified loxP site, a second structure containing a covalent 3′-phosphotyrosine intermediate was obtained. It shows that the ApT step in the top strand is cleaved first. The cleavage induces a 3 Å shift of the C-terminal part of the subunit, containing helices M and N, as well as a redistribution of the kink within the spacer region of the DNA. In contrast to conclusions drawn from a previous structure containing a symmetrized lox site with a distinctly different spacer conformation, we propose that a Cre-induced kink next to the scissile phosphodiester activates the DNA for cleavage. The moving of helices M and N observed in the 3′-phosphotyrosine intermediate could be responsible for the interconversion of Cre subunits through an interaction with the ‘non-cleaving’ subunit, and in particular with its loop 198–208, a major component of the conformational switch.
The interaction between Cre monomers bound to the right and left arms of the loxP recombination site induces a defined bending of the DNA and an anti-parallel arrangement of the DNA duplexes in the synaptic complex and thereby determines which strand will be cleaved first. In an induced-fit mechanism changes in the DNA structure are coupled with conformational changes in the protein and protein–DNA contacts in the spacer region. Tetramerization in the synapse may induce further bending and/or stabilization of the DNA conformation by reciprocal exchange of the C-terminal helix and interactions of N-terminal domains between the Cre subunits bound to different duplexes, followed by coordinated cleavage by the ‘cleavage-competent’ subunits at the ApT sites in the top strands. Apparently, already at the stage of synapsis a conformation of the DNA is induced by the recombinase, which is essentially maintained during the first round of strand cleavage, strand exchange, re-ligation and formation of the HJ intermediate.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the help by Hiang Teo-Dreher in the initial stages of this project and our colleagues from the BW-7B (EMBL Hamburg outstation), ID29 and ID14 beamlines (ESRF, Grenoble), and to Armin Wagner (Swiss Light Source, Viligen) for help with the data collection. This work was supported by a grant from the Volkswagenstiftung to D.S. E.E. is recipient of an EMBO long-term fellowship.
PDB accession nos 1nzb, 1ouq
REFERENCES
- 1.Van Duyne G.D. (2001) A structural view of cre-loxp site-specific recombination. Annu. Rev. Biophys Biomol. Struct., 30, 87–104. [DOI] [PubMed] [Google Scholar]
- 2.Gu H., Marth,J.D., Orban,P.C., Mossmann,H. and Rajewsky,K. (1994) Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science, 265, 103–106. [DOI] [PubMed] [Google Scholar]
- 3.Testa G. and Stewart,A.F. (2000) Creating a transloxation. Engineering interchromosomal translocations in the mouse. EMBO Rep., 1, 120–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchholz F., Refaeli,Y., Trumpp,A. and Bishop,J.M. (2000) Inducible chromosomal translocation of AML1 and ETO genes through Cre/loxP-mediated recombination in the mouse. EMBO Rep., 1, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y. and Bradley,A. (2001) Engineering chromosomal rearrangements in mice. Nat. Rev. Genet., 2, 780–790. [DOI] [PubMed] [Google Scholar]
- 6.Lewandoski M. (2001) Conditional control of gene expression in the mouse. Nat. Rev. Genet., 2, 743–755. [DOI] [PubMed] [Google Scholar]
- 7.Guo F., Gopaul,D.N. and van Duyne,G.D. (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature, 389, 40–46. [DOI] [PubMed] [Google Scholar]
- 8.Gopaul D.N., Guo,F. and Van Duyne,G.D. (1998) Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J., 17, 4175–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo F., Gopaul,D.N. and Van Duyne,G.D. (1999) Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc. Natl Acad. Sci. USA, 96, 7143–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woods K.C., Martin,S.S., Chu,V.C. and Baldwin,E.P. (2001) Quasi-equivalence in site-specific recombinase structure and function: crystal structure and activity of trimeric Cre recombinase bound to a three-way Lox DNA junction. J. Mol. Biol., 313, 49–69. [DOI] [PubMed] [Google Scholar]
- 11.Martin S.S., Pulido,E., Chu,V.C., Lechner,T.S. and Baldwin,E.P. (2002) The order of strand exchanges in Cre-LoxP recombination and its basis suggested by the crystal structure of a Cre-LoxP Holliday junction complex. J. Mol. Biol., 319, 107–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoess R., Wierzbicki,A. and Abremski,K. (1987) Isolation and characterization of intermediates in site-specific recombination. Proc. Natl Acad. Sci. USA, 84, 6840–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoess R., Abremski,K., Irwin,S., Kendall,M. and Mack,A. (1990) DNA specificity of the Cre recombinase resides in the 25 kDa carboxyl domain of the protein. J. Mol. Biol., 216, 873–882. [DOI] [PubMed] [Google Scholar]
- 14.Tribble G., Ahn,Y.T., Lee,J., Dandekar,T. and Jayaram,M. (2000) DNA recognition, strand selectivity and cleavage mode during integrase family site-specific recombination. J. Biol. Chem., 275, 22255–22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee G. and Saito,I. (1998) Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene, 216, 55–65. [DOI] [PubMed] [Google Scholar]
- 16.Lee L. and Sadowski,P.D. (2003) Sequence of the loxP site determines the order of strand exchange by the Cre recombinase. J. Mol. Biol., 326, 397–412. [DOI] [PubMed] [Google Scholar]
- 17.Collaborative Computational Project, Number 4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Cryst. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z. and Minor,W. (1996) In Carter,C.W.,Jr and Sweet,R.M. (eds), Methods in Enzymology. Academic Press, Vol. 276, pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 19.Navaza J. (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr., 57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- 20.de la Fortelle E. and Bricogne,G. (1997) In Carter,C.W.,Jr and Sweet,R.M. (eds), Macromolecular Crystallography. Academic Press, Vol. 276A, pp. 472–494. [Google Scholar]
- 21.Brunger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S., Read,R.J., Rice,L.M., Simonson,T. and Warren,G.L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr., 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 22.Ennifar E., Carpentier,P., Ferrer,J.L., Walter,P. and Dumas,P. (2002) X-ray-induced debromination of nucleic acids at the Br K absorption edge and implications for MAD phasing. Acta Crystallogr. D Biol. Crystallogr., 58, 1262–1268. [DOI] [PubMed] [Google Scholar]
- 23.Marcus Y. (1988) Ionic radii in aqueous solutions. Chem. Rev., 88, 1475–1498. [Google Scholar]
- 24.Eckstein F. (1985) Nucleoside phosphorothioates. Annu. Rev. Biochem., 54, 367–402. [DOI] [PubMed] [Google Scholar]
- 25.Kitts P.A. and Nash,H.A. (1988) An intermediate in the phage lambda site-specific recombination reaction is revealed by phosphorothioate substitution in DNA. Nucleic Acids Res., 16, 6839–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitts P.A. and Nash,H.A. (1988) Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J. Mol. Biol., 204, 95–107. [DOI] [PubMed] [Google Scholar]
- 27.Abremski K. and Hoess,R. (1984) Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J. Biol. Chem., 259, 1509–1514. [PubMed] [Google Scholar]
- 28.McFail-Isom L., Shui,X. and Williams,L.D. (1998) Divalent cations stabilize unstacked conformations of DNA and RNA by interacting with base pi systems. Biochemistry, 37, 17105–17111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.