Abstract
A novel genome vector using the 4215 kb Bacillus subtilis genome provides for precise target cloning and processing of the cloned DNA to the desired structure. Each process highly dependent on homologous recombination in the host B.subtilis is distinguished from the other cloning systems. A 120 kb mouse jumonji (jmj) genomic gene was processed in the genome vector to give a series of truncated sub-megasized DNA. One of these truncated segments containing the first intron was copied in a plasmid by a recombinational transfer method developed for B.subtilis. DNA manipulation previously considered difficult is argued with respect to DNA size and accuracy.
INTRODUCTION
With sequence information now available for various organisms from viruses to higher eukaryotes, functions of not only gene products but also genomic DNA under physiological conditions are drawing much attention. Methods to clone DNA exceeding 100 kb, such as YACs (1) using Saccharomyces cerevisiae and BACs (2) and PACs (3) using Escherichia coli extrachromosomal replicons, were developed to provide DNA libraries in an era when nucleotide sequence information was limited, and successfully contributed to the construction of transgenic mice (4,5) as well as plants (6). Recent sequence-based investigations of large sized DNA (7,8) required rapid and reliable methods. However, the cloning process in these plasmid-born vectors largely depends on restriction endonucleases or physically sheared DNA. In addition, these vectors lack choices for further manipulation of cloned DNA. For example, an elaborate DNA manipulation such as preparation of truncated DNA of various sizes can be achieved partially by inducible homologous recombination in a BAC system using the functions of genes recA (9) or recE and recT (10–12).
A novel cloning and flexible manipulation of sub-megasized DNA was introduced as an alternative approach (13,14). The 4215 kb genome of Bacillus subtilis strain168 (15) plays the role of a cloning vehicle in the system. Direct cloning of target DNA as large as 120 kb (13,14) requires only two flanking segments. These two fragments, called landing pad sequences (LPSs), determine the region to be cloned. Prior to cloning, a set of these LPSs correctly aligned in an E.coli pBR322 plasmid should be pre-installed into a pBR sequence locus of the B.subtilis genome by homologous recombination (Fig. 1A). The aligned LPSs, called the LPS array (LPA), in the genome is the place for actual cloning. A surprisingly high accuracy of homologous recombination (HR) in competent B.subtilis cells mediated integration of the intervening segment between the LPSs (13,14). This HR-mediated precise target cloning is the principle underlying the platform known as the Bacillus subtilis genome (BGM) vector (16).
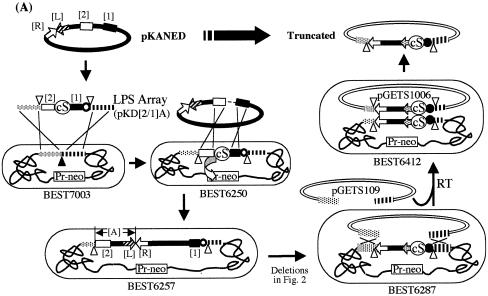
Figure 1.
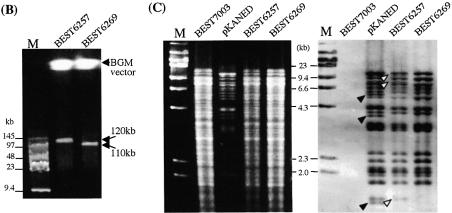
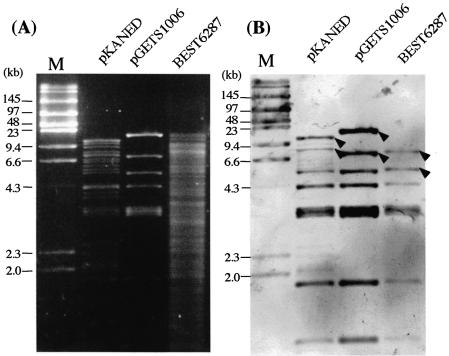
(A) DNA cloning and recovery in the BGM system. BEST7003 acts as a BGM vector. Dotted and striped boxes in the BEST7003 genome indicate pBR322 tet-half and amp-half (13). The BAC vector part of pKANED is represented by two arrows [L] and [R]. The cat gene for E.coli selection is included in the [R]. The LPS array (LPA) is constructed in the pBR322-based plasmid pCISP311B. The positive selection marker, circled cS, is between fragments [1] and [2]. The cS is composed of the cI repressor gene and spectinomycin resistance gene. The LPA part of pKD[2/1]A was delivered in the genomic pBR sequence of BEST7003 by double homologous recombination as indicated by X. In BEST6250, the newly introduced cI repressor gene product binds the Pr promoter of Pr-neo and shuts off expression of the neomycin phosphotransferase gene (neo), rendering the strain sensitive to neomycin. The intervening segment between [1] and [2] of pKANED is integrated into BEST6250 through double crossing-over. Conversion of the insert of BEST6257 to that of BEST6287 is described in Figure 2. Truncated 34 kb of BEST6287 was copied in the pGETS109 vector by recombinational transfer (RT) and purified as pGETS1006. The dotted arrow of BEST6287, BEST6412 and pGETS1006 is described in Figure 2. The closed circle indicates the cat gene for B.subtilis and the open circle indicates the erm gene. The closed triangle indicates the tet gene. The I-PpoI recognition sequence is indicated by an open triangle. (B) Cloned 120 and 110 kb DNA of BEST6257 and BEST6269 digested with I-PpoI and resolved by CHEF. The BGM vector part (4.2 Mb) migrates slowly. Running conditions: 4.67 V cm–1, 30 s pulse time and 16 h running time at 14°C. (C) HindIII digests of genomes of BEST7003, BEST6257 and BEST6269 run by CHEF (left) and hybridized with pKANED as the probe (right). BEST6257 and BEST6269 carry bands derived from pKANED. Bands indicated by closed arrowheads are lost in BEST6257. Bands indicated by open arrowheads in region [A] are deleted in BEST6269. Running condition: 3 V cm–1, 18 s pulse time and 16 h running time at 14°C.
The same principle can be applied to conversion of the cloned DNA in the BGM vector. We adopted a mouse genomic jumonji gene (jmj) spanning 200 kb which maps on chromosome 13 (17–19). As eukaryotic gene expression might be regulated by many factors (20,21), some of which could interact with the large first intron, we focused our investigation on specific protein–DNA interaction with the 90 kb long jmj intron. In this report a protocol is presented for systematic deletion and recovery of the deleted clone as purified DNA.
MATERIALS AND METHODS
Bacterial transformation
Escherichia coli and B.subtilis were grown at 37°C unless specified. Transformation was carried out as described previously (22). The degree of B.subtilis competency was standardized by the conversion of leuB to leuB::tet as described (22). Selection by antibiotics for both E.coli and B.subtilis was done under the conditions reported previously (13). Bacillus subtilis genomic DNA in liquid or in an agarose plug was prepared as described (22).
Bacillus subtilis strain as a BGM vector
All bacterial strains used in this study are listed in Table 1. The B.subtilis strain, BEST7003, was derived from RM125 that had lost a Bsu restriction–modification system (23). Two DNA fragments were inserted into the genome of BEST7003. Briefly, one is a 4.3 kb sequence of an E.coli plasmid pBR322 inserted into the NotI site of the proB gene positioned at 1378 kb (15,24,25), and the other is a Pr promoter-controlled neomycin resistance gene (Pr-neo) inserted between positions 3516 and 3522 kb (24,25). The former genomic pBR322 sequence provides a DNA cloning locus and always remains at both ends of the cloned insert, as shown in Figures 1A and 2. Expression of the latter Pr-neo gene is regulated by the presence or absence of its repressor cI gene (24). This internal selection system is used for the direct selection of recombinants with the combinational use of a cI repressor gene (13,24). The selection scheme from BEST7003 to BEST6257 is shown in Figure 1A.
Table 1. Bacterial strains constructed in this study.
| Bacterial strain | Genotype or insert | Transformation | Antibiotic selectiona | PCR | Reference or source |
|---|---|---|---|---|---|
| Escherichia coli | |||||
| JA221 | F– hsdR hsdM+ trp leu lacY recA1 | (31) | |||
| Bacillus subtilis | |||||
| RM125 | leuB8 arg-15 ΔSPβ | (23) | |||
| BEST7003b | RM125 plus proB::pBRTc Pr-neo | Nm, Tc | This study | ||
| BEST6250 | proB::pKD[2/1]A | pKD[2/1]A × BEST7003 | Spc, Em | This study | |
| BEST6257 | proB::pBR[120] | pKANED × BEST6250 | Nm, Em | P1, P2, P3 | This study |
| BEST6265 | proB::pBR[110] | pCISP320 × BEST6257 | Spc, Em | P1, P2, P3 | This study |
| BEST6269 | proB::pBR[110] | pCISP320B × BEST6265 | Nm, Em | P1, P2, P3 | This study |
| BEST6283 | proB::pBR[86] | pKDC 302 × BEST6269 | Cm, Spc | P1, P2 | This study |
| BEST6287 | proB::pBR[34] | pKDS103 × BEST6269 | Cm, Spc | P1 | This study |
| BEST6280 | proB::pBR[16.5] | pKDC202 × BEST6269 | Cm, Spc | P1 | This study |
| BEST6273 | proB::pBR[11.2] | pKane301 × BEST6269 | Em, Spc | P1 | This study |
| BEST6412 | proB::pBR[34], pGETS1006 | pGETS109 × BEST6287 | Cm, Spc, Tc | P1 | This study |
| BEST6410 | proB::pBR[16.5], pGETS1004 | pGETS109 × BEST6280 | Cm, Spc, Tc | P1 | This study |
| BEST6426 | proB::pBR[11.2], pGETS1011 | pGETS103 × BEST6273 | Em, Spc, Tc | P1 | This study |
aNm, neomycin resistance; Spc, spectinomycin resistance; Cm, chloramphenicol resistance; Em, erythromycin resistance; Tc, tetracycline resistance.
bThe Pr-neo gene is inserted between NotI sites of yvfC and yveP.
Figure 2.
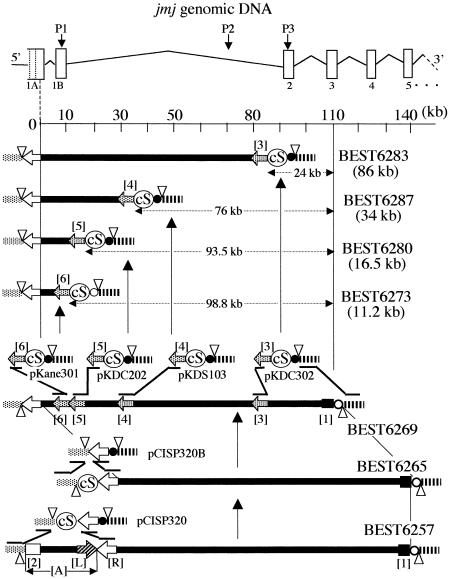
Systematic nested deletion in the BGM vector. The structure of the genomic jmj gene with exons (open rectangles) and introns is presented at the top. Locations of specific PCR primers P1, P2 and P3 are indicated. Only the cloning locus of BEST6257 is shown at the bottom, including the pBR sequence (tet-half and amp-half). The regional specific deletion is caused by plasmids [3] (pKDC302), [4] (pKDS103), [5] (pKDC202) and [6] (pKane301). Segments [3]–[6], not drawn to scale, were separately prepared to carry out deletion. They were linearized using appropriate restriction enzymes in the transformation of BEST6269. Transformants were selected with chloramphenicol. BEST6273 was selected using spectinomycin. Other symbols are as presented in Figure 1.
Bacterial plasmids
All bacterial plasmids used in this study are listed in Table 2. pCISP310B and pCISP311B derived from pBR322 were used to deliver the LPA in the genomic pBR322 sequence of BEST7003. pGETS109 was derived from pGETS103 (26), which shuttles between E.coli and B.subtilis. Plasmids other than the BAC clone were prepared from 50 ml E.coli cultures in the presence of antibiotics for selection and purified using a Plasmid Purification Kit (Qiagen, Tokyo, Japan). BAC clones were prepared from 200 ml cultures and purified with a CsCl gradient formed by ultracentrifugation in the presence of ethidium bromide (27). Plasmids from B.subtilis were purified by a method described in the literature (26). Type II restriction enzymes and T4 DNA ligase were obtained from Toyobo (Tokyo, Japan), except NotI (TaKaRa, Kyoto, Japan) and I-PpoI (Promega, Madison, USA). PCR-mediated amplification was carried out using TaKaRa Ex Taq™ Hot Start Version.
Table 2. Escherichia coli plasmids constructed in this study.
| Plasmid | Construction or featuresa | Reference |
|---|---|---|
| pCISP310B | ApR, SpR, CmR, BSR | (13) |
| pCISP311B | ApR, SpR, EmR, BSR | (13) |
| pBAC108L | BAC vector (6.8 kb), CmR | (13) |
| pKANED | CmR | This study |
| pBAC108Lbsr | bsr-marked pBAC108L | (13) |
| pBR::BAC102 | Chimeric plasmid with pBR322 and pBAC108Lbsr | (13) |
| LPS clones isolated from BamHI library of pKANEDb | ||
| pKane101 | 8.6 kb insert containing a 2 kb LPS [1] | |
| pKane201 | 9.5 kb [2] | |
| pKane301 | 8.7 kb [6] | |
| LPS clones isolated from BamHI library of pKANEDc | ||
| pKDC202 | 2.5 kb [5] | |
| pKDC302 | 5 kb [3] | |
| pKDS103 | 9.5 kb [4] | |
| LPS clone derivatives | ||
| pCISP320 | A 2.3 kb HindIII fragment BAC[R] of pBR::BAC102 cloned into the HindIII site of pCISP310B | |
| pCISP320B | Deletion of [cI] (2.6 kb BamHI fragment) from pCISP320 | |
| LPS array plasmid | ||
| pKD[2/1]A | A 2 kb EcoRI fragment [1] from pKane101 cloned into the EcoRI site of pKane201 | |
| Recovery plasmid | ||
| pGETS103 | The θ-type replicating plasmid of thermophilic Bacillus carrying the pBR322 sequence and tetracycline resistance gene | (24; unpublished) |
| pGETS109 | Derived from pGETS103. | |
| pGETS1006 | Recovery plasmid carrying 34 kb of BEST6287 | |
| pGETS1004 | Recovery plasmid carrying 16.5 kb of BEST6280 | |
| pGETS1011 | Recovery plasmid carrying 11.2 kb of BEST6273 |
aApR, ampicillin resistance; SpR, spectinomycin resistance; CmR, chloramphenicol resistance; BSR, blasticidin S resistance; EmR, erythromycin resistance; [cI], cI cassette.
bThe library was made in the BamHI site of pCISP311B.
cThe library was made in the BamHI site of pCISP310B.
RESULTS
Mouse BAC clone
The sequence of the mouse genomic jmj locus mapped at chromosome 13 (18) was obtained from the public database (http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?org=mouse&chr=13&maps=cntg-r,bes,scan,ugMm,loc&VERBOSE=ON&query=Jmj&cmd=focus&fill=40&size=40&compress=no). A BAC clone pKANED (23908) purchased from Incyte Genomics Inc. was screened using a primer set within the jumonji gene: 5′-GGA AGA GGA AGA AAA GTC GG-3′ and 5′-CCT TGC TCA TTC TGA GAT CC-3′. pKANED had a 140 kb insert that fortuitously contains exons 1A–5. There are putative ESTs in the 90 kb long first intron and they are conserved in the large first intron of the human counterpart found in the database (http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?ORG=hum&chr=6&maps=cntg-r,clone,scan,ugHs,loc&VERBOSE=ON&cmd=focus&fill=40&size-40&query=JMJ).
Cloning in the BGM vector
Cloning of the 120 kb DNA region of pKANED that covers the first and second introns required two DNA fragments, LPS[1] and LPS[2]. LPS[1] and LPS[2] were prepared from a BamHI library of pKANED made in the pBR322 plasmid in order to avoid possible mutations caused during PCR- mediated cloning. These two LPSs were assembled in pCISP311B in the correct order and orientation for cloning the designated 120 kb DNA, resulting in the plasmid pKD[2/1]A. Integration of pKD[2/1]A into the BEST7003 genome generated BEST6250, as shown in Figure 1A.
Transformation of BEST6250 by pKANED generated a recombinant BEST6257 that was selected using neomycin as presented in Figure 1A. Cloning of the 120 kb fragment was confirmed as follows. I-PpoI digestion of the BEST6257 genome produced a 120 kb fragment which was resolved by CHEF electrophoresis (Fig. 1B). The fine structure of the cloned 120 kb DNA was analyzed by Southern hybridization of HindIII digests using pKANED as a probe (Fig. 1C). Southern signals observed in BEST6257 were identical to those originating from pKANED. With additional Southern experiments for EcoRI and BamHI digests (data not shown) it was demonstrated that the cloning process and maintenance of the 120 kb mouse DNA in the BGM vector produced no structural alterations.
The 120 kb DNA of BEST6257 was converted to a 110 kb sequence of BEST6269 carrying the first and second exons only. This processing was conducted as two steps as described in Figure 2. In the first step, the 10 kb [A] segment was replaced with the cI gene and spectinomycin resistance gene (cI-spc) of pCISP320, resulting in BEST6265. In the second step, BEST6269 that had lost the cI-spc part was obtained. Removal of cI-spc was carried out by similar positive selection used to isolate BEST6257.
Systematic deletion formation
To prepare first intron segments of various lengths, the 110 kb fragment was shortened. The introduction of a deletion was performed using homologous recombination by the reverse order of cloning. For example, to obtain 86 kb, the 24 kb beyond fragment [3] of BEST6269 should be deleted as indicated in Figure 2. Plasmid pKDC302 was constructed for this purpose and is listed in Table 2. Homologous recombination of fragment [3] and the pBR sequences (amp-half shown as a striped box in Fig. 2) with their counterpart of pKDC302 caused replacement of the 24 kb plus erm gene by the two antibiotic resistance markers spc and cat of the pKDC302 plasmid. BEST6283 was obtained as a representative of 83 recombinants per µg of pKDC302 selected with chloramphenicol, having lost erythromycin resistance. This method leaves the pBR sequence at both ends, which can be used for recovery as described below.
Similarly, pKDS103, pKDC202 and pKane301 were constructed to delete the 76, 93.5 and 98.8 kb regions beyond the indicated fragments [4], [5] and [6]. Strains carrying truncated DNA, BEST6287 and BEST6280, were obtained as chloramphenical-resistant recombinants of BEST6269 using pKDS103 and pKDC202. BEST6273 was selected with spectinomycin using pKane301. The number of recombinants per µg plasmid DNA selected using chloramphenicol varied from 40 to 352. To our surprise, shortening of sub-megasized DNA in the BGM system was carried out with high efficiency. Sizes of the truncated DNAs (86, 34, 16.5 and 11.2 kb) were determined by I-PpoI digestion as shown in Figure 3A. The fine structure was confirmed by genomic Southern analysis using pKANED as a probe, as presented in Figure 3B. Amplification of the PCR fragments listed in Table 1 further confirmed the presence of these sequences in the recombinants.
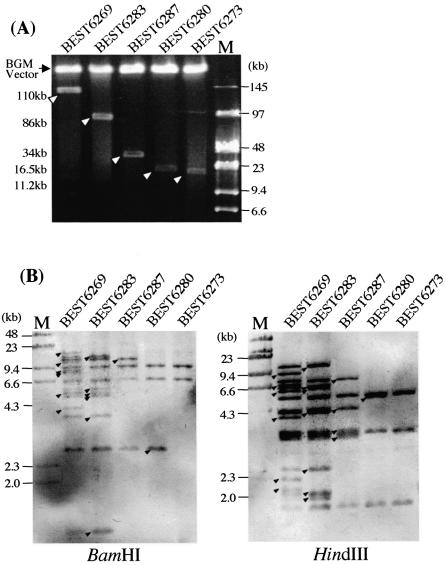
Figure 3.
Shortened sub-megasized DNA in the BGM vector. (A) I-PpoI fragments resolved by CHEF are indicated by open arrowheads with their sizes on the left. The BGM vector (4.2 Mb) migrates slowly. Running conditions: 4 V cm–1, 6 s pulse time and 20 h running time at 14°C. (B) Genomic DNA of indicated strains digested by BamHI (left) and HindIII (right) were run by CHEF: 3 V cm–1, 6 s (for BamHI digestion) or 18 s (for HindIII digestion) pulse time and 16 h running time at 14°C. Southern bands detected using pKANED as a probe are consistent with those predicted from the determined sequence. Closed arrowheads indicate fragments lost by deletion.
Recovery from the BGM vector by recombinational transfer
Because we are interested in protein factors that interact within the long first intron, the truncated DNAs of BEST6287, BEST6280 and BEST6273 would be suitable as probes for screening. Isolation of the I-PpoI fragment from an agarose gel suffers from unnecessary physical shearing, low yield and inability for further amplification. We have recently established a method to recover the DNA cloned in the BGM vector as a covalently closed circular form. According to the method described in Tsuge and Itaya (26), the insert DNA flanked by two pBR sequences (amp-half and tet-half) was copied in the pGETS103 plasmid based on the recombinational transfer mechanism (Fig. 1A).
We used pGETS109 derived from pGETS103 to recover the 34 kb segment of BEST6287. The copy number of pGETS109 remains as low as 1 per cell, which avoids unnecessary interference between plasmids (manuscript in preparation). The transformant selected with tetracycline should harbor the pGETS1006 plasmid repaired by copying the 34 kb, because linearized pGETS109 never circularizes by itself (see Fig. 1A). Four tetracycline-resistant transformants were obtained from BEST6287 using pGETS109 linearized with HindIII. Faithful copying of the genomic 34 kb segment in the isolated pGETS1006 plasmid was confirmed by Southern blot analysis as shown in Figure 4A and B. The two altered HindIII fragments from pGETS1006 were assigned to the two junctions of pGETS109, indicating conversion of the 34 kb from genomic to plasmid DNA. The pGETS1006 plasmid was immediately transferred to the E.coli host by the nature of the pBR322 replicon of pGETS109. Similarly, the truncated 16.5 and 11.2 kb fragments were obtained as pGETS1004 and pGETS1011 (data not shown). pGETS1006 is used for the reverse genetics of jmj with the DNA originating from strain129/SVJ.
Figure 4.
Recovered DNA from the BGM vector. (A) DNA of the indicated plasmids (pKANED and pGETS1006) and of the BEST6287 genome were digested by HindIII and resolved by CHEF. Running conditions: 2.67 V cm–1, 18 s pulse time and 16 h running time at 14°C. (B) Southern analysis using pGETS1006 as a probe identified bands transferred from BEST6287 to pGETS1006. Two bands indicated by closed arrowheads were caused by junction to different vectors.
DISCUSSION
A novel BGM system was introduced for the preparation of large DNA suitable for investigation of transcriptional regulation. The BGM system has the potential to provide properly manipulated DNA starting from the direct cloning of sub-megasized DNA, through to conversion of the cloned DNA to the desired sequence, and its recovery as covalently closed circular (ccc)DNA for further use. One of the most important characteristics of the homologous recombination-mediated system is the flexibility to convert the cloned DNA. Structures of all cloned DNAs, including the four truncated versions, were stably maintained even when the recombinants were cultured in non-selective medium. The stability likely comes from the fact that the cloned DNA is replicated as part of the B.subtilis genome, which is consistent with our previous reports (13–15).
Combined with our previous results (13,26) and unpublished observation that other BAC clones of up to 200 kb were successfully cloned, the findings of this study prove that DNA of ∼100 kb can be routinely cloned into the BGM vector. As the elongation of cloned inserts was examined previously (13), the capacity of the BGM vector to carry far larger DNA needs to be investigated. The length of the LPS in which homologous recombination proceeds affects the efficiency of cloning as well as deletion. As the LPS employed in this study generated sufficient numbers of recombinants, we expect that a longer LPS would be more effective. An appropriate combination of LPS length and clonable DNA size may allow direct cloning from mouse genomic DNA.
In addition to cloning, the deletion proceeded with surprisingly high efficiency. The BGM system is superior with respect to accuracy, compared with other inducible systems such as the recA-mediated recombination process (9) or the GET recombination system (10–12). These E.coli systems require either the counterselection step in the recA-dependent homologous recombination (9) or elimination of selection markers by the site-specific recombinase FLP (28) or Cre (29). The manipulation leaves small scars in the final DNA structure, such as FLP target recognition sites (FRT) or loxP sites. In our BGM system the use of more elaborate manipulations such as insertion (14) and inversion (30) allows DNA conversion without leaving flaws in the DNA. Improvements for high throughput use of the BGM system are now being made based on PCR technology. As B.subtilis does not discriminate between DNA sequences in the integration process, the expeditious manipulation of sub-megasized inserts of all BAC clones is expected. A similar handling of DNA from different sources is in progress. It may be advantageous that cloned and manipulated DNA can be stored at room temperature in long-lasting spore forms of B.subtilis (14).
REFERENCES
- 1.Burke D.T., Carle,G.F. and Olson,M.V. (1987) Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science, 236, 806–812. [DOI] [PubMed] [Google Scholar]
- 2.Shizuya H., Birren,B., Kim,U.J., Mancino,V., Slepak,T., Tachiiri,Y. and Simon,M. (1992) Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA, 89, 8794–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pierce J. and Sternberg,N.L. (1992) Using bacteriophage P1 system to clone high molecular weight genomic DNA. Methods Enzymol. 216, 549–574. [DOI] [PubMed] [Google Scholar]
- 4.Strauss W.M., Dausman,J., Beard,C., Johnson,C., Lawrence,J.B. and Jaenisch,R. (1993) Germ line transmission of a yeast artificial chromosome spanning the murine alpha1(I) collagen locus. Science, 259, 1904–1907. [DOI] [PubMed] [Google Scholar]
- 5.Schedl A.M.L., Kelsey,G. and Schutz,G. (1993) A yeast artificial chromosome covering the tyrosinase gene confers copy number-dependent expression in transgenic mice. Nature, 362, 258–261. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton C.M. (1997) A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene, 200, 107–116. [DOI] [PubMed] [Google Scholar]
- 7.Lander E.S., Linton,M.L., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W., Funke,R. et al., The International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 8.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A., Gocayne,J.D. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 9.Yang X.W., Model,P. and Heintz,N. (1997) Homologous recombination based modification in Escherichia coli and germline transmission in transgenenic mice of a bacterial artificial chromosome. Nat. Biotechnol., 15, 859–865. [DOI] [PubMed] [Google Scholar]
- 10.Mikhail N., Robert,W. and Panayiotis,A.I. (2000) Insertion of disease-causing mutation in BACs by homologous recombination in Escherichia coli. Nucleic Acids Res., 28, e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michael O., Mikhail,N., Jim,V., Faten,Z., Robert,W. and Panayiotis,A.I. (2000) Engineering EGFP reporter constructs into a 200 kb human β-globin BAC clone using GET Recombination. Nucleic Acids Res., 28, e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narayanan K., Williamson,R., Zhang,Y., Stewart,A.F. and Ioannou,P.A. (1999) Efficient and precise engineering of a 200 kb beta-globin human/bacterial artificial chromosome in E. coli DH10B using an inducible homologous recombination system. Gene Ther., 6, 442–447. [DOI] [PubMed] [Google Scholar]
- 13.Itaya M., Nagata,T., Shiroishi,T., Fujita,K. and Tsuge,K. (2000) Efficient cloning and engineering of giant DNAs in a novel Bacillus subtilis genome vector. J. Biochem., 128, 869–875. [DOI] [PubMed] [Google Scholar]
- 14.Itaya M. (1995) Toward a bacterial genome technology: integration of the Escherichia coli prophage lambda genome into the Bacillus subtilis 168 chromosome. Mol. Gen. Genet., 248, 9–16. [DOI] [PubMed] [Google Scholar]
- 15.Itaya M. (1993) Integration of repeated sequences (pBR322) in the Bacillus subtilis 168 chromosome without affecting the genome structure. Mol. Gen. Genet., 241, 287–297. [DOI] [PubMed] [Google Scholar]
- 16.Dubnau D. (1999) DNA uptake in bacteria. Annu. Rev. Microbiol., 53, 217–244. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi T., Yamazaki,Y., Katoh-Fukui,Y., Tsuchiya,R., Kondo,S., Motoyama,J. and Higashinakagawa,T. (1995) Gene trap capture of a novel mouse gene jumonji, required for neural tube formation. Genes Dev., 9, 1211–1222. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi T. (1997) jumomji gene maps to mouse chromosome 13. Genomics, 45, 240–242. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi T., Kojima,M., Nakajima,K. and Kondo,S. (1999) jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech. Dev., 86, 29–38. [DOI] [PubMed] [Google Scholar]
- 20.Atchison M.L. (1988) Enhancers: mechanisms of action and cell specificity. Annu. Rev. Cell. Biol., 4, 127–153. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen T., Sherratt,P.J. and Pickett,C.B. (2003) Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol Toxicol., 43, 233–260. [DOI] [PubMed] [Google Scholar]
- 22.Itaya M. (1999) Genetic transfer of large DNA inserts to designated loci of the Bacillus subtilis 168 genome. J. Bacteriol., 181, 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itaya M. and Tanaka,T. (1997) Predicted and unsuspected alterations of the genome structure of genetically defined Bacillus subtilis 168 strains. Biosci. Biotechnol. Biochem., 61, 56–64. [Google Scholar]
- 24.Itaya M. (1999) Effective cloning of unmarked DNA fragments in the Bacillus subtilis 168 genome. Biosci. Biotechnol. Biochem., 63, 602–604. [DOI] [PubMed] [Google Scholar]
- 25.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessières,P., Bolotin,A., Borchert,S. et al. (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 26.Tsuge K. and Itaya,M. (2001) Recombinational transfer of 100-kilobase genomic DNA to plasmid in Bacillus subtilis 168. J. Bacteriol., 183, 5453–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Cherepanov P.P. and Wackernagel,W. (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene, 158, 9–14. [DOI] [PubMed] [Google Scholar]
- 29.Heiss R.H., Zeise,M. and Sternberg,N. (1982) P1 site-specific recombination—nucleotide sequence of the recombining sites. Proc. Natl Acad. Sci. USA, 79, 3398–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toda T., Tanaka,T. and Itaya,M. (1996) A method to invert DNA segments of the Bacillus subtilis 168 genome by recombination between two homologous sequences. Biosci. Biotechnol. Biochem., 60, 773–778. [DOI] [PubMed] [Google Scholar]
- 31.Itaya M. (1990) Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc. Natl Acad. Sci. USA, 87, 8587–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]