Abstract
In plants, cell-to-cell transport of endogenous and viral proteins and ribonucleoprotein complexes (RNPCs) occurs via plasmodesmata. Specificity of this transport pathway appears to involve interaction between such proteins/RNPCs and plasmodesmal chaperones/receptors. Here, KN1 and the cucumber mosaic virus movement protein (CMV-MP) were used, in a modified phage-display screening system, to identify peptides capable of interacting with proteins present in a plasmodesmal-enriched cell wall fraction. Binding/competition assays and microinjection experiments revealed that these phage-displayed peptides and homologous synthetic oligopeptides function as ligand-specific antagonists of macromolecular trafficking through plasmodesmata. A KN1 peptide antagonist had the capacity to interact with a motif involved in the dilation of plasmodesmal microchannels. Although KN1 could still achieve limited movement through plasmodesmata when this SEL motif was blocked, KN1-mediated transport of KN1–sense RNA was fully inhibited. These findings provide direct support for the hypothesis that KN1 requires, minimally, two physically separated signal motifs involved in the dilation of, and protein translocation through, plasmodesmal microchannels, and provide direct proof that plasmodesmal dilation is a prerequisite for the cell-to-cell transport of an RNPC.
Keywords: peptide antagonist/phage display/plasmodesmal transport of macromolecules/SEL motif
Introduction
One strategy of plant cells to overcome the challenge of intercellular communication through their intervening cellulose walls was to develop cytoplasmic channels. The nature of these structures, termed plasmodesmata, has been the focus of considerable attention in recent years (Lucas, 1995, 1999; Mezitt and Lucas, 1996; Ghoshroy et al., 1997; McLean et al., 1997; Ding, 1998; Ding et al., 1999; Lazarowitz and Beachy, 1999). The plasmodesmal supramolecular complex establishes a specialized form of cytoplasmic continuity, comprised of an outer plasma membrane and a centrally located appressed endoplasmic reticulum (ER), which also forms a continuum with the ER of the adjacent cells. Proteins embedded within the plasma membrane and appressed ER extend into the cytoplasmic annulus, thereby dividing this pathway into a number of microchannels having diameters of the order of 2.5–3.0 nm (Ding, 1998). Intercellular exchange of metabolites occurs through these plasmodesmal microchannels (Robards and Lucas, 1990).
Plasmodesmata also mediate the transport of macromolecules, including endogenous and viral-encoded proteins and ribonucleoprotein complexes (RNPCs). Mutant analyses performed on such proteins have underscored the specificity of protein–plasmodesmal interaction (Fujiwara et al., 1993; Noueiry et al., 1994; Waigmann et al., 1994; Lucas et al., 1995; Rojas et al., 1997; Ishiwatari et al., 1998; Lough et al., 1998). In addition, this plasmodesmal intercellular transport system appears to share features in common with intracellular translocation mechanisms in which the transfer of macromolecules occurs in either a folded or unfolded state (Subramani, 1996; Kragler et al., 1998; Schatz, 1998). In this regard, numerous studies have established that during protein and RNPC translocation, the size exclusion limit (SEL) of the plasmodesmal microchannels undergoes a significant increase. In the quiescent state, in which macromolecules are not being trafficked, the plasmodesmal SEL is <1 kDa (Terry and Robards, 1987; Wolf et al., 1989), whereas when proteins and/or RNPCs are being translocated the SEL increases to values ranging from 10 to 50 kDa, depending on the protein and/or tissue(s) involved (Wolf et al., 1989; Fujiwara et al., 1993; Lucas et al., 1995; Waigmann and Zambryski, 1995; Balachandran et al., 1997; Rojas et al., 1997; Imlau et al., 1999; Oparka et al., 1999). This measured increase in plasmodesmal SEL could be a prerequisite for, or a consequence of, RNPC translocation; i.e. dilation of the plasmodesmal orifice may be essential to permit the entry and transport of a large cargo, such as an RNPC.
Mechanistic aspects of plasmodesmal-mediated protein translocation have been examined using KNOTTED1 (KN1), a maize homeobox transcription factor (Jackson et al., 1994; Lucas et al., 1995). Microinjection experiments conducted with structurally stabilized KN1 provided support for the hypothesis that KN1 undergoes partial unfolding during translocation through the plasmodesmal microchannel; however, the KN1-induced increase in plasmodesmal SEL does not require protein unfolding (Kragler et al., 1998). Based on these studies, cell-to-cell transport of proteins (and RNPCs) via plasmodesmata appears to involve three events: (i) recognition of a plasmodesmal translocation signal sequence by a putative specific chaperone/receptor; (ii) induction of an increase in plasmodesmal SEL; and (iii) unfolding and translocation through the dilated microchannel. As a corollary to this model, it can be hypothesized that proteins which undergo cell-to-cell movement display two signals, mediating (i) the SEL increase and (ii) translocation through plasmodesmata.
A powerful approach for the identification of small protein motifs capable of interacting with receptors is afforded via phage-display assays. This approach has also been developed to identify potential antagonists that can interfere with receptor–ligand interactions (Doorbar and Winter, 1994; Goodson et al., 1994; Szardenings et al., 1997). To advance our understanding of the role played by microchannel dilation (SEL increase) in protein translocation, this phage-display approach was employed in an effort to isolate peptides capable of interfering with this specific step in cell-to-cell transport. To this end, we used a plasmodesmal-enriched cell wall fraction (W2 fraction proteins), previously shown to contain proteins involved in cell-to-cell movement (Kragler et al., 1998). As the phage display was originally developed for use with highly purified interaction partners, but the W2 fraction contains an array of proteins, we modified the assay to allow the identification of interaction motifs present within such a heterogeneous mixture. This new assay resulted in the isolation of specific oligopeptides capable of interacting with putative chaperones/receptors involved in the cell-to-cell trafficking of proteins and RNPCs. Furthermore, characterization of enriched phage 12mer peptides and synthetic oligopeptides, using in vitro binding/competition assays and microinjection experiments, identified specific motifs capable of selectively interfering with the cell-to-cell transport of macromolecules. A peptide reflecting a motif located within the N-terminal region of KN1 acted as a powerful antagonist against the ability of KN1 to potentiate an increase in plasmodesmal SEL. Even though KN1 could still achieve a limited degree of movement through plasmodesmata, when this SEL motif was blocked, KN1-mediated transport of KN1–sense RNA was fully inhibited. These findings provide the first direct evidence that protein-mediated dilation of plasmodesmata is a prerequisite for the cell-to-cell transport of an RNPC.
Results
Oligopeptides capable of interacting with plasmodesmal proteins are identified by screening a random phage library
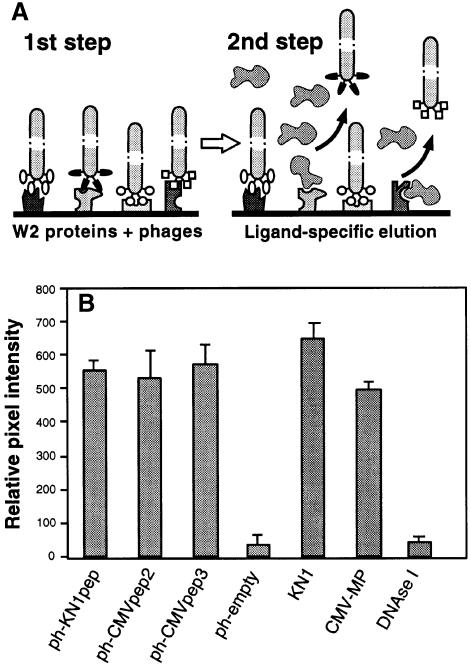
A random M13 phage library, exposing 12mer amino acid sequences in the context of the minor coat protein, was employed in a screen to isolate oligopeptides capable of interacting with putative plasmodesmal proteins contained within a tobacco W2 cell fraction (Kragler et al., 1998). As certain endogenous and viral movement proteins interact competitively with plasmodesmal components involved in cell-to-cell transport of macromolecules, specificity of the oligopeptide interaction was probed using KN1 and cucumber mosaic virus movement protein (CMV-MP). In these experiments, aliquots of W2 fraction proteins were dotted onto nitrocellulose membrane strips that were then incubated with blocking buffer containing 1% (w/v) bovine serum albumin (BSA) to saturate non-specific interactions. The immobilized W2 fraction proteins were then incubated with a suspension of the phage library, with subsequent elution of specific bound phages being achieved by exposure to excess levels of KN1 or CMV-MP (see Figure 2A). As a control for this elution step, an Escherichia coli cell lysate was used as the eluent, since this represented the background in which the KN1 and CMV-MP were expressed. Isolated phages were amplified and the incubation/elution steps repeated four times. Finally, after five panning rounds, 16 individual phages were selected at random from each of the three elution reactions; ssDNA encoding each modified phage was then sequenced for further analysis.
Fig. 2. Isolation and characterization of phages displaying random 12mer peptides that interact with proteins contained within a plasmodesmal-enriched W2 cell fraction. (A) Schematic representation of the protocol employed to screen for peptides capable of interacting with putative plasmodesmal receptors contained within the W2 cell fraction prepared from N.tabacum (tobacco) plants. Immobilized W2 proteins were first incubated with the phage peptide library. After extensive washing to remove unbound phages, ligand competition was carried out to displace a specific subset of the bound phages (ligand-specific elution). Note that non-specific binding of this ligand, to W2 protein-bound phages, would not cause their release. (B) Binding activity between isolated phage-displayed peptides and W2 fraction proteins. Phage probes were first coated onto 6 nm gold particles (ph-KN1pep, ph-CMVpep2, ph-CMVpep3 and ph-empty) and then used for in vitro binding assays to test for their interaction with putative plasmodesmal proteins contained in the W2 cell fraction. KN1-gold, CMV-MP-gold and DNase I-gold probes were employed as controls. Extracted W2 fraction protein (100 ng) was probed with 200 ng of phage-gold, KN1-gold, CMV-MP-gold or DNase I-gold complexes. Interaction of protein-gold probes was visualized by silver enhancement. Data were compiled from three to four replicates, for each experiment, and represent relative pixel densities [after subtraction of background as measured with BSA-gold (see Kragler et al., 1998)]; mean ± SEM.
As shown in Figure 1A, elution of phages by addition of KN1 led to the 100% enrichment of a specific epitope, in that the 16 phages obtained from these experiments exposed the same sequence of amino acids (oligopeptide termed KN1pep). The relative probability that KN1pep would be present 16 times within the phage library was calculated to be 6 × 10–97. Furthermore, the likelihood of identifying this peptide, 16 times, by sequencing randomly selected phages after panning, is represented by an even smaller number! Hence, the identity in the amino acid composition of the isolated peptide-displaying phages must reflect a high specific enrichment of an epitope that is able to bind to a plasmodesmal constituent (putative chaperone/receptor) present within the tobacco W2 cell fraction. In equivalent elution experiments, in which CMV-MP was employed, a more diverse array of oligopeptides was identified (Figure 1A; CMVpep1–4). However, significant enrichment was obtained, in that CMVpep1 and CMVpep2 were represented in seven and six phages, respectively. As with KN1pep, the relative probability that these peptides occur in the phage library at this frequency is 1 × 10–41 and 5 × 10–32, respectively. Again, the chance of finding these phage peptide sequences in randomly selected phages is zero. In the CMV-MP screen, one phage of the 16 analyzed exposed no 12mer peptide. Control elution experiments carried out with an E.coli cell lysate yielded 15 different oligopeptides. An additional ‘empty’ phage was also isolated, which did not express a 12 amino acid peptide. None of these control oligopeptides exhibited any significant homology in their primary structure to KN1pep or CMVpep1, -2, -3 or -4 (data not shown).
Fig. 1. Isolation and sequence analysis of plasmodesmal-interacting peptides. (A) A phage-display library was used in a screen to identify peptides that interacted with proteins in a plasmodesmal-enriched Nicotiana tabacum cell wall fraction (W2 fraction proteins). Proline residues have been indicated in bold type and basic residues are highlighted. (B) Amino acid sequence homology between identified phage-displayed 12mer peptides and motifs within the KN1 and CMV-MP. The bold type KN1 sequence represents the 14 amino acid oligopeptide, KN1-Npepsynth, used in microinjection experiments. (Single-letter code used for amino acid residues.)
Candidate protein motifs identified by comparison with enriched phage oligopeptide sequences
Oligopeptides obtained from the elution of the phages with KN1 and CMV-MP were compared with the amino acid sequences of these same proteins. To investigate the significance of these peptide sequences we first compared the 15 random 12mer peptides, isolated using the E.coli lysate, against KN1 and CMV-MP. This control established a median homology of 45 ± 8%; this represents the inherent chance of a random peptide sequence aligning within KN1 or CMV-MP. As illustrated in Figure 1B, KN1pep and the identified KN1 motif exhibited ∼75% homology. Based on these analyses, the epitope reflected in KN1pep was identified in a structural motif extending from amino acid residues 35 to 47 of the KN1-deduced amino acid sequence. In addition, motifs homologous to KN1pep were not detected in the CMV-MP at a homology score above that of the random control.
The epitope reflected in CMVpep1 appeared to reside in a CMV-MP motif located in the region from amino acid residue 237 to 246 (Figure 1B; 90% homology). The immediate neighboring region of the CMV-MP, from residue 224 to 233, and the overlapping region from residue 240 to 248, appeared to reflect the CMVpep2 epitope (80 and 66% homology, respectively). Although no motifs homologous to CMVpep3 were detected in the CMV-MP, two possible motifs reflecting the CMVpep4 epitope were identified in the region of residues 20–31 (92% homology) and residues 225–234 (70% homology). This analysis of the CMV-MP strongly implicated two structural motifs, one in the region of codon 20–31 and the second from codon 224 to 248, which are related to CMVpep1, -2 and -4.
Identified oligopeptides were next analyzed in terms of their predicted structural and physical properties. All oligopeptides reflected a general hydrophilic character, as deduced from calculations of the hydrophobicity index (Kyte and Doolittle, 1982). Despite the fact that residues such as arginine and cysteine are known to interfere with the minor coat protein III, in terms of it mediating phage penetration into E.coli cells (Peters et al., 1994), charged arginine residues were present in displayed peptides of KN1pep, CMVpep1, -2 and -3.
In vitro binding/competition studies confirm that specific phage-displayed oligopeptides interact with W2 fraction proteins
As representative oligopeptides, KN1pep, CMVpep2 and CMVpep3 were selected for further analysis to establish their capacity to interact, in vitro, with putative plasmodesmal proteins contained within the tobacco W2 cell fraction. Phages exposing these specific oligopeptides were isolated, coated onto 6 nm gold particles and used in dot-blot interaction assays (Kragler et al., 1998). The intensity of the signal associated with these phage-gold probes was visualized by silver enhancement and measured using the NIH Image software. As illustrated in Figure 2B, these three phages were able to interact with immobilized W2 fraction proteins. Indeed, the phage-gold probes displayed binding activities equivalent to those obtained with either KN1-gold or CMV-MP-gold; these values were >10-fold higher than those recorded using an empty phage (control; ph-empty) that did not expose a 12mer peptide. Additional experiments conducted with DNase I-gold added further support for the hypothesis that the exposed KN1pep, CMVpep2 and CMVpep3 mediate an interaction with the W2 fraction proteins (Figure 2B).
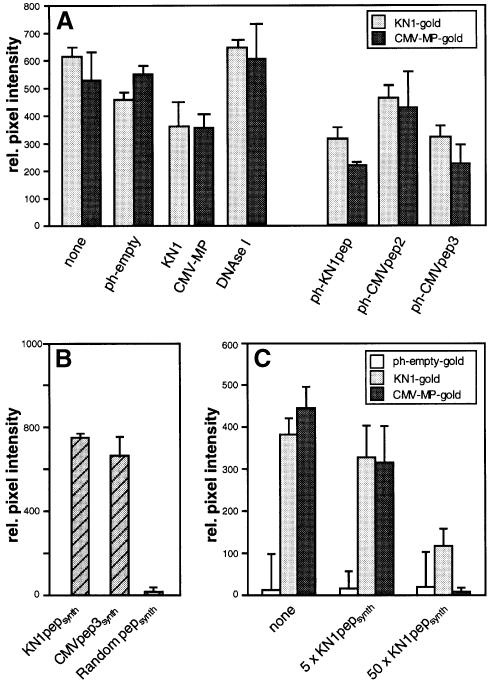
Specificity of this phage 12mer peptide-mediated interaction with the W2 fraction proteins was next investigated. Here, competition assays were conducted, using both KN1 and CMV-MP, under conditions reflecting those existing in the original screen (Figure 2A). For these studies, KN1-gold or CMV-MP-gold, and specific phage 12mer peptides were present in near equimolar amounts. Due to the size and physical properties of the phages, higher concentrations were found to result in the precipitation of KN1-gold and CMV-MP-gold probes; unfortunately, this precluded the performance of concentration-dependent competition assays. Control competition experiments were first performed with ph-empty, KN1 or CMV-MP probes bound to W2 fraction proteins. The presence of ph-empty had little effect on KN1-gold and CMV-MP-gold binding to the W2 fraction proteins (Figure 3A). As shown previously (Kragler et al., 1998), the presence of an equimolar amount of KN1/CMV-MP reduced the binding of the gold probe by ∼30%, in contrast to DNase I, which did not interfere (compete) with the binding of the gold probes.
Fig. 3. Specificity of binding between isolated phage peptides and the tobacco W2 fraction proteins. (A) Competitive binding interactions between isolated tobacco W2 fraction proteins and KN1, CMV-MP and specific phage-displayed 12mer peptides. KN1 and CMV-MP were coated onto 6 nm gold particles and then used to determine specific protein interactions with the W2 cell fraction. Immobilized W2 fraction proteins (100 ng) were first incubated with the indicated probes (equimolar to the gold probe) and then subsequently competed with KN1-gold or CMV-MP-gold (0.2 µg). Note that experiments depicted on the left represent controls performed under the same conditions as described above for the phage peptide probes. (B) Ability of KN1pepsynth, CMVpep3synth and random pepsynth gold probes (0.2 µg) to bind immobilized W2 fraction proteins. (C) Competitive binding between KN1-gold, CMV-MP-gold or ph-empty-gold (all present at 0.2 µg) and a 5× or 50× molar excess of KN1pepsynth. Immobilized W2 fraction proteins were incubated with a mixture of competitor and gold probe. Interaction of protein-gold probes was visualized by silver enhancement. Data were compiled from three to four replicates for each experiment, and represent relative pixel densities (after subtraction of background as measured with BSA-gold); mean ± SEM.
The capacity of KN1-gold or CMV-MP-gold to displace the bound phages from the W2 fraction proteins was greater for CMVpep2 (ph-CMVpep2) and lower for KN1pep (ph-KN1pep) and CMVpep3 (ph-CMVpep3). These studies revealed that the phage peptides, KN1-gold and CMV-MP-gold probes interacted competitively for the same W2 fraction protein(s). Although no sequence homology was detected between CMVpep3 and CMV-MP, ph-CMVpep3 acted in a manner similar to ph-KN1pep; thus, it appears to represent a synthetic ligand. Consistent with this notion, ph-KN1pep and ph-CMVpep3 appeared to interact with the W2 fraction proteins with affinities comparable to KN1 or CMV-MP (Figure 3A).
In view of the fact that increasing amounts of phage particles initiated aggregation of gold probes, we next performed concentration-dependent competition assays using synthetic KN1pep (KN1pepsynth) and CMVpep3 (CMVpep3synth) oligopeptides as probes. Initial control binding experiments, conducted using a random 14mer oligopeptide gold-labeled probe, established that such short peptides per se do not bind to the W2 fraction proteins (Figure 3B). In marked contrast, both KN1pepsynth and CMVpep3synth probes bound to the W2 fraction proteins in a manner similar to KN1 and CMV-MP. As shown in Figure 3C, increasing the level of KN1pepsynth resulted in a marked displacement of both KN1-gold and CMV-MP-gold probes. Equivalent results were obtained when similar competition experiments were conducted with a synthetic CMVpep3 (CMVpep3synth) probe (data not shown). These results established that KN1pepsynth and CMVpep3synth interact, competitively, with KN1 and CMV-MP for binding to W2 fraction proteins.
Microinjection experiments establish that phage-displayed oligopeptides interact in vivo with plasmodesmal proteins
Phages displaying the peptides KN1pep, CMVpep2 and CMVpep3 were next employed in microinjection experiments to ascertain whether they have the capacity to interact in vivo with the putative plasmodesmal interaction partners contained within the W2 cell fraction. Control injections were first performed on mesophyll cells to ensure that the interconnecting plasmodesmata were functioning in a normal manner. As shown in Table I, both KN1-fluorescein isothiocyanate (FITC) and KN1 plus 11 kDa F-dextran (FITC-labeled dextran) displayed efficient movement from the target cell into the surrounding cells of the mesophyll tissue. Consistent with our previous studies (Kragler et al., 1998), efficient cell-to-cell movement was also obtained when either CMV-MP or the W2 fraction proteins were coinjected with 11 kDa F-dextran. Note that introduction of the small, membrane-impermeant fluorescent probe, Lucifer yellow CH, into a target cell resulted in the rapid and extensive cell-to-cell movement of this probe, thereby establishing that plasmodesmata in this field of cells permitted the diffusion of small molecules. In addition, when 11 kDa F-dextran was injected alone, the fluorescent signal was almost always confined to the target cell; in the few cases where the signal did move, it was restricted to the adjacent cells. In these infrequent cases, at the time of injection, either cell-to-cell transport of endogenous macromolecules was likely to have been taking place, or the plasmodesmal microchannels were in the processes of closure, following such transport. In any event, this value served as a baseline against which to compare the cell-to-cell transport of microinjected probes.
Table I. Influence of specific phages on cell-to-cell movement of endogenous and viral movement proteins when coinjected into mesophyll cells of fully expanded leaves of Nicotiana benthamiana.
| Injected probe | Coinjected agent | Injections | Movement (%) |
|---|---|---|---|
| Lucifer yellow CH | – | 45 | 44 (98)a |
| 11 kDa FITC-dextran | – | 55 | 3 (5)b |
| KN1 | F-dextranc | 48 | 39 (81)a |
| CMV-MP | F-dextran | 18 | 15 (83)a |
| W2 proteins | F-dextran | 21 | 19 (90)a |
| KN1-FITC | – | 33 | 29 (88)a |
| ph-KN1pepd | F-dextran | 37 | 1 (3)b |
| ph-KN1pepe | KN1 + F-dextran | 39 | 9 (23)f |
| ph-KN1pep | W2 proteins + F-dextran | 30 | 9 (30)f |
| ph-KN1pep | KN1-FITC | 57 | 31 (54)g |
| ph-KN1pep | CMV-MP + F-dextran | 30 | 17 (57)h |
| ph-CMVpep2d | F-dextran | 15 | 2 (13)b |
| ph-CMVpep2 | KN1 + F-dextran | 30 | 6 (20)f |
| ph-CMVpep2 | W2 proteins + F-dextran | 25 | 3 (12)f |
| ph-CMVpep2 | KN1-FITC | 26 | 20 (77)g |
| ph-CMVpep2 | CMV-MP + F-dextran | 30 | 6 (20)f |
| ph-CMVpep3d | F-dextran | 22 | 4 (18)b |
| ph-CMVpep3 | KN1 + F-dextran | 35 | 19 (54)h |
| ph-CMVpep3 | W2 proteins + F-dextran | 23 | 18 (78)a |
| ph-CMVpep3 | KN1-FITC | 31 | 21 (68)a |
| ph-CMVpep3 | CMV-MP + F-dextran | 35 | 13 (37)f |
| ph-KN1pep | KN1 + CMV-MP + F-dextran | 14 | 4 (29)f |
| ph-CMVpep3 | KN1 + CMV-MP + F-dextran | 13 | 5 (38)f |
| ph-empty | KN1 + CMV-MP + F-dextran | 23 | 18 (78)a |
| ph-emptyd | F-dextran | 20 | 3 (15)b |
| ph-empty | KN1 + F-dextran | 41 | 27 (65)h |
| ph-empty | KN1-FITC | 15 | 11 (73)a |
| ph-empty | W2 proteins + F-dextran | 23 | 18 (78)a |
| ph-empty | CMV-MP + F-dextran | 28 | 22 (79)a |
aFluorescent signal associated with the reporter probe moved through >10–15 surrounding mesophyll cells.
bWhen movement was detected, the fluorescent signal was generally limited to adjacent cells.
cCoinjection of 11 kDa FITC-labeled dextran (F-dextran) allowed monitoring of plasmodesmal SEL increase and cell-to-cell movement of unlabeled KN1, W2 fraction proteins or CMV-MP.
dConcentration of various phage particles used in microinjection experiments was 1 µg/µl.
eRatio of phage particles to KN1, W2 fraction proteins or CMV-MP was ∼1:4 (w/w).
fWhen the fluorescent signal moved it did so slowly and was mostly restricted to the adjacent cell(s).
gRate of KN1-FITC movement was much slower relative to the controls.
hF-dextran signal was detected in 5–10 surrounding mesophyll cells.
Delivery into mesophyll cells of ph-KN1pep, ph-CMVpep2, ph-CMVpep3 or ph-empty, along with 11 kDa F-dextran, resulted in the confinement of the fluorescent probe to the injected cell (Table I). As in the 11 kDa F-dextran control experiments, on the few occasions when fluorescence was detected beyond the injected cell, it was again confined to the adjacent cells. These experiments established that none of these oligopeptides has the capacity either to induce an increase in plasmodesmal SEL or to mediate the cell-to-cell transport of the associated phage. In addition, based on confocal laser scanning microscopy (CLSM) imaging of injected mesophyll cells, introduction of these phages did not cause any detectable changes in general subcellular structure (data not shown).
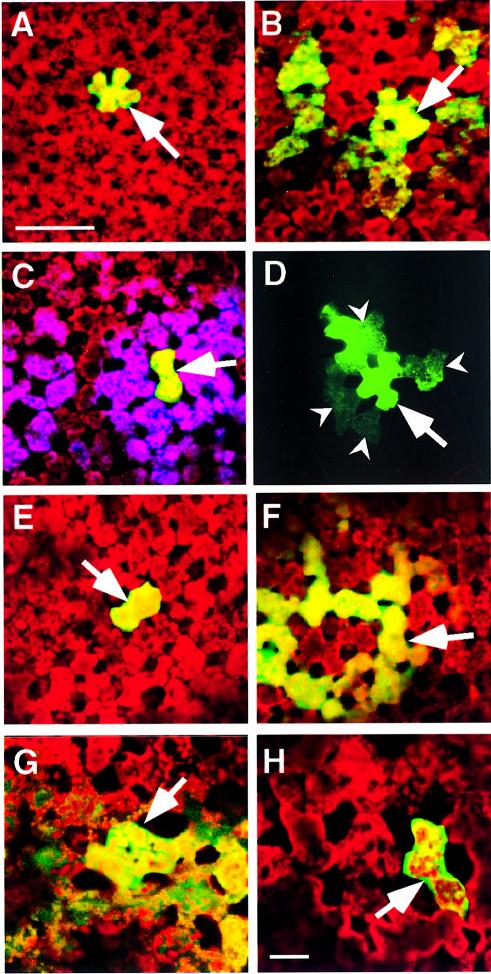
To ascertain whether the KN1pep epitope had the capacity to interact with plasmodesmal components involved in KN1 transport, a series of coinjection experiments was performed with ph-KN1pep and KN1. As illustrated in Figure 4A and Table I, in the presence of ph-KN1pep, the ability of KN1 to increase plasmodesmal SEL and mediate its own cell-to-cell transport was dramatically inhibited. Control injection experiments, in which KN1 was delivered along with ph-empty (Figure 4B; Table I), indicated that the presence of ph-empty had a detectable effect on KN1 movement; however, compared with ph-KN1pep, the extent of this inhibitory interaction appeared to be minimal. In addition, the possibility of non-specific blockage of plasmodesmata by such treatments was discounted by double-microinjection experiments. In the first injection, the 11 kDa F-dextran introduced along with ph-KN1pep and KN1 remained within the target cell. A second injection was then used to deliver Lucifer yellow CH into this same cell, and the fluorescent signal associated with this probe was always (six out of six experiments) observed to move quickly and extensively throughout the neighboring mesophyll (Figure 4C).
Fig. 4. Phage-displayed peptides and synthetic peptides act as antagonists to KN1-induced increase in plasmodesmal SEL and cell-to-cell movement of macromolecules. All microinjection experiments were carried out on mesophyll cells located within fully expanded leaves of N.benthamiana. A Leica confocal laser scanning microscope was used to simultaneously collect fluorescent signals in the FITC (green) and chlorophyll (red) channels, 10 min after probes were delivered into a target cell. Optical sections, collected in each channel, were stacked and then combined to generate the images presented. (A) Coinjection of ph-KN1pep, KN1 and 11 kDa F-dextran; fluorescent signal (F-dextran) was confined to the target mesophyll cell. (B) Coinjection of ph-empty, KN1 and 11 kDa F-dextran; note that the green fluorescent signal moved from the target cell into neighboring mesophyll cells. (C) Double microinjection experiment; first injection as in (A), in the second injection, Lucifer yellow CH (purple false color) was delivered into the same target cell and moved into the adjacent cells and beyond. (D) KN1-FITC and ph-KN1pep coinjected into a target cell; fluorescent signal associated with KN1 was detected in adjacent cells (darts). Red autofluorescence has been omitted from this image to enhance the visualization of the KN1-FITC signal. (E) Coinjection of KN1pepsynth, KN1 and 11 kDa F-dextran; fluorescent signal (F-dextran) was confined to the target mesophyll cell. (F) Coinjection of CMVpep3synth, KN1 and 11 kDa F-dextran; fluorescent signal (F-dextran) detected in the surrounding mesophyll cells. (G) KN1-mediated transport of KN1–sense RNA-CF. (H) KN1-mediated transport of KN1–sense RNA-CF is blocked in the presence of KN1pepsynth. Arrows indicate the injected cells; scale bar in (A) = 100 µm, and is common to (B)–(F); scale bar in (H) = 50 µm, and is common to (G).
As the ph-KN1pep was isolated by elution from the W2 cell fraction, using KN1, a parallel series of microinjection experiments was next performed with W2 fraction proteins. Here, equivalent results were obtained, in that ph-empty had only a small inhibitory effect on the W2 fraction protein-mediated cell-to-cell movement of 11 kDa F-dextran, whereas this capacity was greatly reduced in the presence of ph-KN1pep (Table I). Interestingly, coinjection of ph-KN1pep along with CMV-MP resulted in a significantly smaller reduction in the ability of CMV-MP to induce an increase in plasmodesmal SEL and mediate its own cell-to-cell transport (Table I). In a reciprocal experiment, ph-CMVpep3 was observed to have only a limited effect, if any, on the capacity of KN1 and W2 fraction protein(s) to interact with plasmodesmata; i.e. 54 and 78% movement, respectively, compared with 65 and 78% obtained in the ph-empty controls (Table I). However, ph-CMVpep3 had a greater inhibitory effect on the capacity of CMV-MP to potentiate the cell-to-cell movement of the coinjected 11 kDa F-dextran (37% movement compared with 79% in the ph-empty controls; Table I). Thus, the KN1pep 12mer exposed by ph-KN1pep has the capacity to specifically block the motifs on KN1 and W2 fraction protein(s) that lead, either directly or indirectly, to dilation of the plasmodesmal microchannels.
Additional experiments in which ph-CMVpep2 was coinjected with KN1, W2 fraction proteins or CMV-MP, established that this oligopeptide had a significant inhibitory effect on the ability of these endogenous and viral movement proteins to potentiate the cell-to-cell movement of the 11 kDa F-dextran (Table I). This suggested that the CMVpep2 epitope interacts with a component of the macromolecular trafficking pathway that is common to a number of endogenous and viral movement proteins. Collectively, these microinjection results support the hypothesis that the competitors, KN1 and CMV-MP, used in the screening of the random phage library, led to the enrichment of phages displaying oligopeptides that can interact specifically with W2 fraction plasmodesmal components involved in protein recognition and delivery to, or transport through, the plasmodesmal microchannel.
Phage-displayed oligopeptides retard but do not prevent protein transport through plasmodesmata
As ph-KN1pep prevented the KN1-mediated movement of 11 kDa F-dextran, we next tested whether the cell-to-cell transport of FITC-labeled KN1 was similarly inhibited by the presence of the ph-KN1pep. As illustrated by the data presented in Table I, coinjection of ph-KN1pep and KN1-FITC did not block the movement of fluorescent signal beyond the target cell. However, compared with control injections involving ph-empty and KN1-FITC, the efficacy of KN1-FITC trafficking was lower (54% movement versus 73% in control ph-empty injections) and the extent of movement was greatly reduced; in most cases, KN1-FITC was restricted to the adjacent cells (Figure 4D). In addition, the rate at which fluorescence associated with KN1-FITC accumulated in these adjacent cells was considerably slower, occurring over many minutes compared with seconds in control experiments.
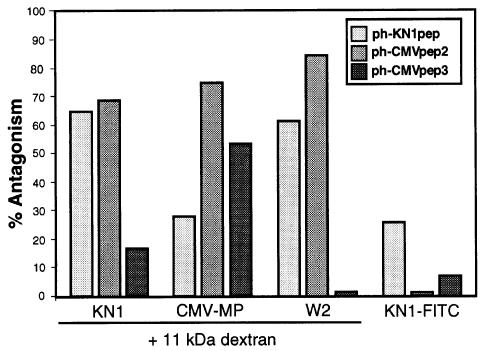
Consistent with the small effect of ph-CMVpep3 on the KN1-mediated movement of 11 kDa F-dextran, coinjection of ph-CMVpep3 and KN1-FITC had no significant effect on the efficacy, extent or rate of movement of fluorescence from the target cell into the surrounding mesophyll (Table I). An equivalent result was obtained in experiments in which KN1-FITC was coinjected with ph-CMVpep2. However, although the efficacy of KN1-FITC transport was high (77% movement), the actual rate was slower when compared with the ph-empty controls. This latter result is highly significant for, as with ph-KN1pep, the ph-CMVpep2 acted as a powerful antagonist against the KN1-mediated movement of 11 kDa F-dextran (Table I). The relative ability of ph-KN1pep, ph-CMVpep2 and ph-CMVpep3 to function as an antagonist against KN1, CMV-MP and the W2 fraction proteins, in terms of their capacity to block the increase in plasmodesmal SEL, is depicted in Figure 5. Here, we also illustrate that these three phages have a low ability to act as antagonists against the cell-to-cell trafficking of KN1-FITC. Collectively, these results provide irrefutable support for the hypothesis that specific oligopeptide-carrying phages can function as antagonists towards KN1, W2 fraction protein(s) and CMV-MP motifs involved (directly or indirectly) in dilating plasmodesmal microchannels (normally observed as an increase in SEL).
Fig. 5. Antagonistic effect of phage peptides on both the ability of KN1, CMV-MP and W2 fraction proteins to increase plasmodesmal SEL and the capacity of KN1-FITC to move cell to cell. Histograms represent the percentage inhibition of movement observed in experiments in which 11 kDa F-dextran was coinjected with KN1, CMV-MP or W2 fraction proteins and ph-CMVpep2, ph-CMVpep3 and KN1pep, respectively. Limited antagonism between the phage peptides and KN1-FITC movement is illustrated by the histograms presented on the right. [The % antagonism values were calculated relative to the percentage cell-to-cell movement obtained in ph-empty control experiments (see Table I).]
KN1 and CMV-MP compete for a common component in the plasmodesmal transport pathway
Recent studies conducted with gold-conjugated KN1 and CMV-MP provided evidence consistent with the hypothesis that these two proteins compete for a common component in the plasmodesmal macromolecular trafficking pathway (Kragler et al., 1998). Experiments in which ph-KN1pep, KN1, CMV-MP and 11 kDa F-dextran were all coinjected into a mesophyll cell provided additional evidence consistent with this hypothesis. Here, cell-to-cell movement (29%) reflected the inhibitory situation observed when ph-KN1pep was coinjected with KN1 plus 11 kDa F-dextran (23% movement), rather than the 57% movement detected when ph-KN1pep was introduced along with CMV-MP and 11 kDa F-dextran. Similarly, introduction of ph-CMVpep3, along with KN1, CMV-MP and 11 kDa F-dextran, resulted in movement of the fluorescent signal in 38% of the injections, which was identical to that observed when ph-CMVpep3 was injected with CMV-MP (37% movement). This value is lower than the 54% movement detected in the presence of ph-CMVpep3, KN1 and 11 kDa F-dextran (Table I). A control for these experiments, in which ph-empty was substituted for ph-KN1pep and ph-CMVpep3, confirmed that these significant reductions in cell-to-cell movement were not attributable to the presence of the phage particles per se. The simplest interpretation of the inhibitory effect of ph-KN1pep on the cell-to-cell movement of CMV-MP, and of ph-CMVpep3 on KN1, is that each antagonist–movement protein combination interferes with a common constituent(s) utilized by KN1 and CMV-MP.
Synthetic KN1 oligopeptides function as antagonists against KN1 motif involved in modulating plasmodesmal SEL
Specificity associated with the in vivo interaction between the ph-KN1pep and the plasmodesmal constituent, involved in microchannel dilation, underlies the potential use of this oligopeptide in the dissection of the molecular events involved in the transport of macromolecules through plasmodesmata. To test this specificity further, in the absence of the phage particle, oligopeptides equivalent to those represented by KN1pep, CMVpep2 and CMVpep3 (i.e. KN1pepsynth, CMVpep2synth and CMVpep3synth) were used in microinjection experiments.
In control studies, coinjection of KN1pepsynth along with 11 kDa F-dextran had little or no effect on plasmodesmal properties as, in most experiments, the fluorescent signal was confined to the injected cell; however, in some cases, a fluorescent signal was detected in the cells adjacent to the target cell (Table II). When coinjected with KN1 and 11 kDa F-dextran, KN1pepsynth was highly effective at blocking the KN1-mediated increase in plasmodesmal SEL (Figure 4E); limited movement of the fluorescent signal was observed in only seven out of 38 (18%) injection experiments, compared with 11 out of 13 (85%) for the KN1 controls (Table II). In these competition experiments, KN1pepsynth was used at an ∼10-fold molar excess over KN1; this molar ratio was found to be effective for in vitro competitive binding assays (Figure 3C).
Table II. Antagonistic effects of synthetic oligopeptides on the capacity of KN1 and CMV-MP to interact with mesophyll plasmodesmata located within fully expanded leaves of Nicotiana benthamiana.
| Injected probe | Coinjected agent | Injections | Movement (%) |
|---|---|---|---|
| Lucifer yellow CH | – | 18 | 17 (94)a |
| 11 kDa FITC-dextran | – | 40 | 7 (18)b |
| KN1 | F-dextranc | 13 | 11 (85)a |
| W2 proteins | F-dextran | 10 | 8 (80)a |
| CMV-MP | F-dextran | 10 | 8 (80)a |
| KN1-FITC | – | 22 | 18 (82)a |
| KN1pepsynth | F-dextran | 37 | 13 (35)b |
| KN1pepsynthd | KN1 + F-dextran | 38 | 7 (18)b |
| KN1pepsynth | KN1-FITC | 39 | 23 (59)e |
| CMVpep3synth | F-dextran | 17 | 3 (18)b |
| CMVpep3synthd | CMV-MP + F-dextran | 14 | 7 (50)e |
| CMVpep3synth | KN1 + F-dextran | 19 | 12 (63)f |
| CMVpep3synth | KN1-FITC | 18 | 15 (83)a |
| KN1-Npepsynth | F-dextran | 20 | 5 (25)b |
| KN1-Npepsynthd | KN1 + F-dextran | 20 | 3 (15)b |
| KN1-Npepsynth | W2 proteins + F-dextran | 22 | 3 (14)b |
| KN1-Npepsynth | KN1-FITC | 20 | 5 (25)b |
| KN1-Npepsynth | CMV-MP + F-dextran | 22 | 18 (70)a |
a–cAs in Table I.
dRatio of synthetic peptide to protein was 3:1 (w/w).
eWhen the fluorescent signal moved it did so slowly and was mostly restricted to the adjacent cell(s).
fF-dextran signal was detected in 5–10 surrounding mesophyll cells.
The inhibition of KN1 function by KN1pepsynth was equivalent to that observed in ph-KN1pep experiments (Tables I and II). Further confirmation of the equivalence in the effects of KN1pepsynth and ph-KN1pep was obtained from studies involving KN1-FITC. As previously observed with ph-KN1pep, KN1pepsynth reduced the extent and rate of KN1-FITC cell-to-cell movement. Again, in the cases where the fluorescent signal was observed to move from the injected cell (59%), it was most often confined to adjacent cells.
To ensure that KN1pepsynth did not cause the closure of plasmodesmal microchannels, the following consecutive injections were performed. First, KN1pepsynth, KN1 and 11 kDa F-dextran were introduced into a target mesophyll cell. As expected, no fluorescence associated with the F-dextran was observed beyond the adjacent cell(s). Next, Lucifer yellow CH was injected into the fluorescently labeled target cell; this small fluorescent probe was observed to move out rapidly into the neighboring cells. This experiment was repeated in triplicate, with the same results being obtained each time. Hence, KN1pepsynth per se does not appear to cause a perturbation to the plasmodesmal microchannels, thus allowing the mesophyll cells to maintain symplasmic continuity.
The level of homology between KN1pep and a 14 amino acid long segment at the N-terminus of KN1 (Figure 1B) suggested that this region may well contain a signal sequence (motif) that mediates the interaction with components of the plasmodesmal translocation machinery. A synthetic oligopeptide, KN1-Npepsynth, consisting of these 14 KN1 residues, was prepared and used in microinjection experiments. Coinjection of KN1-Npepsynth along with KN1 resulted in the dramatic inhibition of 11 kDa F-dextran movement (Table II). An equivalent result was obtained in experiments involving KN1-Npepsynth and W2 fraction proteins. A noteworthy result was obtained when KN1-FITC was coinjected with KN1-Npepsynth; under these conditions the capacity of KN1-FITC to move out from the injected cell was greatly reduced. A comparison between KN1pepsynth and KN1-Npepsynth experiments highlights this result, in that cell-to-cell movement of KN1-FITC occurred in 59 and 25% of the injections, respectively. Importantly, KN1-Npepsynth had little or no effect on the capacity of CMV-MP to interact with plasmodesmata, further underscoring the specificity of the interaction between KN1, KN1-Npepsynth and the plasmodesmal component(s).
Our attempts to dissolve CMVpep2synth in protein buffer were unsuccessful and thus, microinjection experiments could not be performed with this peptide. Microinjection experiments conducted with CMVpep3synth revealed that it had only a slight inhibitory effect on the ability of KN1 to mediate cell-to-cell movement of 11 kDa F-dextran (Figure 4F). Furthermore, when coinjected with KN1-FITC, CMVpep3synth did not inhibit KN1-FITC movement (Table II). However, when coinjected with CMV-MP plus 11 kDa F-dextran, the CMVpep3synth restricted the movement of fluorescence signal to the adjacent cells. These results further confirmed the specificity of the competitive interaction between KN1pepsynth and KN1; in addition, they established that CMVpep3synth can be used as an internal control in future experiments.
Increase in plasmodesmal SEL is essential for KN1-mediated transport of KN1–sense RNA
Earlier studies demonstrated that KN1 potentiates the selective cell-to-cell transport of its own sense RNA (Lucas et al., 1995). Control experiments employing covalently labeled chromatide-fluorescein RNA (KN1–sense RNA-CF) confirmed this finding (Figure 4G; Table III). The converse experiment in which KN1–sense RNA was coinjected with KN1-FITC yielded identical results. These experiments confirmed that KN1 mediates the cell-to-cell transport of its own KN1–sense RNA, presumably in the form of an RNPC. As ph-KN1pep and KN1pepsynth blocked the KN1-mediated increase in plasmodesmal SEL, these probes were next used to determine whether KN1-potentiated trafficking of KN1–sense RNA could occur in the absence of an increase in SEL. Coinjection of ph-empty (control), along with KN1-FITC and KN1–sense RNA, had only a slight effect on the movement of the fluorescently labeled KN1-FITC (73% movement compared with 83% in the absence of ph-empty). However, coinjection of ph-KN1pep with KN1-FITC and KN1–sense RNA resulted in a dramatic reduction in the cell-to-cell movement of the KN1-FITC fluorescent signal (Table III). This result indicated that an increase in SEL is likely to be a prerequisite for the cell-to-cell movement of the KN1–RNA complex. This hypothesis was further tested by microinjection experiments involving KN1pepsynth and KN1-Npepsynth. As a control for these synthetic peptide studies, KN1 and KN1–sense RNA-CF were coinjected with CMVpep3synth; as expected, normal movement of the CF signal was observed under these conditions (Table III). In marked contrast, inclusion in the injection mixture of either KN1pepsynth or KN1-Npepsynth dramatically reduced the cell-to-cell movement of the RNPC to 10 and 16%, respectively (Figure 4H; Table III). Clearly, these results provide support for the hypothesis that peptide antagonists can alter the functional properties of plasmodesmata.
Table III. Synthetic KN1 oligopeptides block KN1-mediated cell-to-cell transport of KN1–sense RNA.
| Injected probea | Coinjected agent | Injections | Movement (%) |
|---|---|---|---|
| Lucifer yellow CH | – | 24 | 24 (100)b |
| 11 kDa F-dextran | – | 23 | 3 (13)b |
| KN1 | F-dextranc | 25 | 21 (84)b |
| KN1-FITC | – | 10 | 9 (90)b |
| KN1–sense RNA-CFd | KN1 | 21 | 17 (81)b |
| KN1–sense RNAe | KN1-FITC | 6 | 5 (83)b |
| KN1–sense RNA | KN1-FITC + ph-empty | 11 | 8 (73)b |
| KN1–sense RNA | KN1-FITC + ph-KN1pep | 9 | 2 (22)f |
| KN1–sense RNA-CF | KN1 + CMVpep3synth | 10 | 7 (70)b |
| KN1–sense RNA-CF | KN1 + KN1pepsynth | 21 | 2 (10)f |
| KN1–sense RNA-CF | KN1 + KN1-Npepsynth | 18 | 3 (16)f |
aAll probes were injected into mesophyll cells located in fully expanded leaves of N.benthamiana.
bFluorescent signal associated with the reporter probe moved through >10–15 surrounding mesophyll cells.
cCoinjection of 11 kDa F-dextran allowed monitoring of plasmodesmal SEL increase and cell-to-cell movement of unlabeled KN1.
dConcentrations of KN1–sense RNA (unlabeled and covalently labeled with CF), KN1/KN1-FITC and KN1pepsynth/KN1-Npepsynth/CMVpepsynth used in these experiments were 10 ng/µl, 2.5 µg/µl and 1.0 µg/µl, respectively.
eConcentrations of KN1–sense RNA, KN1/KN1-FITC and ph-KN1pep/ph-empty used in these experiments were 10 ng/µl, 2.5 µg/µl and 1.0 µg/µl, respectively.
fWhen movement was detected, the fluorescent signal was generally limited to adjacent cells.
Discussion
An ever increasing number of plant- and viral-encoded proteins have been shown to move from cell to cell via plasmodesmata. However, few of the genes encoding proteins that form this intercellular trafficking machinery have been characterized. As chaperones and plasmodesmal receptor/docking molecules are likely to be involved in the delivery of these endogenous movement proteins (Mezitt and Lucas, 1996) and RNPCs (Kragler et al., 1998; Lee et al., 2000), a method was sought for the identification of these plasmodesmal constituents. To this end, an in vitro screen was developed, based on a 12mer random peptide phage-display library, which allowed for the identification of putative motifs, within KN1 and the CMV-MP, that interact with proteins involved in the cell-to-cell trafficking of macromolecules. The foundation for these experiments was provided by earlier studies in which we demonstrated that both KN1 and CMV-MP interact specifically with proteins contained within a plasmodesmal-enriched cell wall fraction [W2 fraction proteins (Kragler et al., 1998)].
The feasibility of the phage screen was verified both by the 12mer peptide enrichment results (Figure 1) and the in vitro binding assays (Figures 2 and 3). The high level of enrichment obtained using the plant-encoded KN1, to competitively displace the bound phages, supports the hypothesis that KN1, and the eluted ph-KN1pep, interacts with a W2 fraction protein(s) in a highly specific manner. The lower enrichment of 12mer peptides in the CMV-MP experiments may simply reflect the presence of a higher number of potential interaction partners within the W2 fraction. It is also important to note that the ability of some of the identified peptides (KN1pep, CMVpep2 and CMVpep3) to interact, in vivo, with components of the plasmodesmal system essential for macromolecular trafficking was established using microinjection methods. Thus, these 12mer peptides should now provide an effective bait for the identification and characterization of KN1 and CMV-MP interaction partners (chaperones and plasmodesmal receptors). Furthermore, the screening method developed in this study should have broad utility in terms of identifying both specific domains/motifs within viral proteins (replicase, coat protein, movement protein, etc.) and the host interacting proteins involved in infection/resistance.
Significant homology was detected between KN1pep and a short N-terminal region of KN1 (Figure 1). A sequence comparison performed between this region of KN1 and other members of the KN homeobox gene family (Jackson et al., 1994) revealed that this motif is not highly conserved, except for very closely related genes [Class I knotted-like proteins (Reiser et al., 2000)]. This most likely reflects the presence of a motif conserved at the secondary structural level; i.e. similar to the situation observed for the nuclear export signal sequences (Bogerd et al., 1996). Comparisons made between CMVpep2 and CMVpep4 and the movement proteins encoded by other members of the cucumovirus family revealed that amino acid residues 225 and 226 of CMV-MP are conserved. Deletion of this region of the cucumovirus MP (i.e. the domain reflected in CMVpep2 and CMVpep4) disables the cell-to-cell spread of the virus (Kaplan et al., 1995). Interestingly, this same region was identified by Melcher (1990) as being homologous to a domain, in the C-terminal region of the tobacco mosaic virus MP, previously shown to be necessary for SEL increase, cell-to-cell movement and virus infection (Berna et al., 1991; Gafny et al., 1992; Waigmann et al., 1994). Furthermore, CMVpep4 is highly homologous to an additional N-terminal region (Figure 1B; codons 20–31) known to be essential for CMV-MP function in viral infection (Kaplan et al., 1995). The amino acid residues of CMVpep4 that are identical to those in the related domain of the CMV-MP are highly conserved between most members of the cucumovirus family. Such sequence comparisons support the contention that the phage peptides identified in the present study resemble motifs present within functional domains of viral movement proteins.
In the present study, we describe the first isolation of antagonists that act to prevent in vivo the protein-induced increase in plasmodesmal SEL. Microinjection studies performed on Nicotiana benthamiana mesophyll cells established the level of specificity associated with the interaction between KN1pep, CMVpep2 or CMVpep3 and the component(s) involved in mediating an increase in plasmodesmal SEL. First, ph-KN1pep acts as a selective antagonist against KN1, or W2 fraction proteins, in blocking the normal increase in SEL (Table I; Figure 5). Here, it is important to note that ph-KN1pep had a markedly reduced inhibitory effect on the ability of CMV-MP to induce an increase in SEL. Secondly, whereas CMVpep3 also reduced the ability of CMV-MP to induce an increase in plasmodesmal SEL, it had only a weak effect on the capacity of KN1 and W2 fraction proteins to dilate plasmodesmata (Figure 5). These findings support the notion of a specific, competitive interaction between KN1pep/KN1 or CMVpep3/CMV-MP and a component of the plasmodesmal trafficking pathway. Thirdly, in contrast, ph-CMVpep2 appears to act non-selectively in that it functions as a strong antagonist against KN1, CMV-MP and W2 fraction proteins (Table I; Figure 5). Thus, this antagonist must interact with a component in common to many endogenous and viral movement proteins.
Control experiments carried out with phages not displaying 12mer peptides (ph-empty), as well as our studies with synthetic peptides (KN1pepsynth, KN1-Npepsynth, CMVpep3synth and random pepsynth) further confirmed the selective nature of the interaction between the identified 12mer peptides and the plasmodesmal protein(s). Coinjection of ph-empty particles with either W2 fraction proteins or CMV-MP did not interfere with the ability of these proteins to dilate the plasmodesmal microchannels (Table I). Interestingly, the presence of ph-empty particles did cause a minor reduction in the ability of KN1 to dilate the plasmodesmata. This small effect of the ph-empty phage particles on KN1 is probably non-specific in nature, as coinjection of KN1 with KN1pepsynth, KN1-Npepsynth or ph-KN1pep yielded an equivalent inhibition of the KN1 capacity to mediate the cell-to-cell movement of 11 kDa F-dextran (Tables I and II). Furthermore, given the similarity between the inhibitory effects of ph-KN1pep and KN1-Npepsynth, on both KN1 and W2 fraction proteins, in terms of SEL increase, it seems probable that this sequence reflects a structural motif present in a number of endogenous movement proteins. Finally, as ph-KN1pep and KN1-Npepsynth had only weak inhibitory effects on the capacity of CMV-MP to increase SEL, these antagonists must either be displaced by CMV-MP, or this viral movement protein bypasses this block via an alternative component (either chaperone or receptor) of the plasmodesmal pathway.
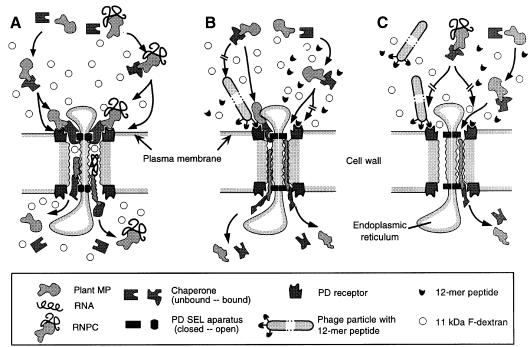
In a previous study we demonstrated that proteins bound to gold particles (6 and 15 nm diameter) are highly effective at blocking the cell-to-cell transport of unbound probes, such as KN1, W2 fraction proteins, CMV-MP and tobacco mosaic virus movement protein (Kragler et al., 1998). This inhibitory effect was attributed to either steric hindrance, due to the size of the gold-protein complexes, or high-affinity binding between a plasmodesmal receptor and the gold-protein complex; both mechanisms would block the cell-to-cell trafficking of macromolecules. In this regard, it is interesting to note that M13 phage particles are comprised of a cylindrical protein shell having a diameter of ∼6 nm and a length of 890 nm. Hence, the diameter of the M13 phage particle is comparable to the 6 nm gold particles used for KN1-gold microinjection experiments and, thus, might well act in an analogous manner to inhibit the function of KN1, CMV-MP or the W2 fraction proteins. However, careful inspection of the microinjection data presented in Table I indicates that a physical block to the entrance of the plasmodesmal microchannel is unlikely to be involved, as in every case where the specific 12mer peptide (i.e. ph-KN1pep, ph-CMVpep2 and ph-CMVpep3) blocked the increase in SEL, KN1-FITC could still move into the adjacent cell(s). Based on these results, it is clear that these peptides do not interact, directly, with KN1 or CMV-MP, as a KN1-FITC molecule bound to a phage would not be able to passage through a plasmodesmal microchannel. Thus, we propose that the phages carrying KN1pep, CMVpep2 and CMVpep3 all interact with receptors that are located either in the orifice of the plasmodesma or are associated with the recognition and translocation of proteins to the plasmodesma (Figure 6).
Fig. 6. Model illustrating the manner in which 12mer peptide-displaying phage particles, or synthetic oligopeptides, interact with the constituents of the plasmodesmal macromolecular (endogenous MP/RNPC) translocation system. (A) Control scenario in which an endogenous MP/RNPC binds to its cognate chaperone for delivery to the plasmodesmal receptor; binding fully activates the SEL motif on the chaperone, leading to dilation of the microchannel. The chaperone–endogenous MP/RNPC then engages the translocation machinery for transport along the length of the microchannel and eventual release into the cytoplasm of the adjacent cell. Note that, during each translocation event, 11 kDa F-dextran molecules enter the microchannel, by diffusion, and are eventually released into the adjacent cell. (B) Ligand-specific competition between the 12mer peptide-displaying phage particles, or the synthetic oligopeptide, and the endogenous MP, for the plasmodesmal receptor site, inhibits the normal docking and microchannel dilation steps; under these conditions, although retarded, the chaperone–endogenous MP can still engage the translocation machinery. With the SEL apparatus in the closed condition, 11 kDa F-dextran cannot enter the microchannel and the endogenous MP must undergo more extensive unfolding to allow movement along the microchannel; upon release into the cytoplasm of the adjacent cell, both the chaperone and the endogenous MP are inactive due to misfolding. (C) Phage-displayed peptides and synthetic oligopeptides prevent the chaperone–RNPC activation of the SEL apparatus, which blocks further transport of the RNPC. Under these conditions, chaperone–endogenous MP behaves as in (B).
The phage-carrying and synthetic peptide antagonists provided a powerful tool to explore further the molecular events underlying protein and RNPC translocation through plasmodesmata. As indicated earlier, ph-KN1pep, ph-CMVpep2, KN1pepsynth and KN1-Npepsynth all blocked the movement protein-mediated increase in SEL. However, in the presence of either ph-KN1pep or KN1pepsynth, KN1-FITC was still able to move through the undilated plasmodesmal microchannel (Tables I and II). These results provide direct proof that cell-to-cell trafficking of proteins can occur in the absence of the normal increase in plasmodesmal SEL. The slowness with which this transport process occurred, combined with the limited extent to which the fluorescently labeled KN1 moved (Figure 4D) probably reflects: (i) steric hindrance imposed on KN1 at the orifice of the undilated microchannel; and (ii) structural changes imposed on KN1 caused during its passage through these constricted microchannels (Figure 6B). The present findings also demonstrate, through the use of small peptide antagonists, that plasmodesmal dilation and protein translocation can be uncoupled (Figure 6). Finally, the requirements for RNPC transport, through the plasmodesmal microchannel, were also revealed using the SEL peptide antagonists ph-KN1pep, KN1pepsynth and KN1-Npepsynth. Although KN1-FITC could still be translocated into the adjacent cell, in the presence of these SEL antagonists, RNPC movement was blocked (Table III; Figure 4H). This establishes that, at least for the KN1–sense RNA RNPC, microchannel dilation is a prerequisite for the cell-to-cell movement of RNA in the form of an RNPC (Figure 6C).
The antagonistic activity of KN1pep and KN1-Npepsynth on endogenous movement protein-mediated increase in plasmodesmal SEL and RNPC movement suggests that such ligand-specific peptides may have utility in terms of analyzing the function of endogenous RNPC trafficking, as well as serving to interdict viral spread, through their expression in transgenic plants. Here, the duly identified ligand(s) could be either improved over corresponding natural protein ligands (e.g. plant proteins that act to block the transport of endogenous movement proteins) or artificial mimotopes (Katz, 1997; i.e. peptides exposing structural or harboring physical properties recognized by a plasmodesmal chaperone/receptor). In any event, as the 12 amino acids of the KN1pep (and the KN1-Npepsynth) display all of the information that is necessary and sufficient to interfere with the KN1-induced increase in plasmodesmal SEL, this ligand-specific probe will next be used to identify the protein(s) involved in this interaction.
Materials and methods
Protein expression and phage display
Plant (KN1) and viral (CMV-MP 3a) movement proteins were expressed in and extracted from E.coli, as described previously (Lucas et al., 1995; Kragler et al., 1998). All protein expression, isolation and purification steps were confirmed by SDS–PAGE and western blot analysis. The phosphate-buffered saline buffer was exchanged, using P6 size exclusion chromatography columns (Bio-Rad, Hercules, CA), to protein buffer [1 mM Tris pH 7.5, 10% (v/v) glycerol, 100 mM NaCl, 1 mM EDTA], followed by protein concentration using micro-concentrators (Microcon-10; Amicon, Beverly, MA) to yield a final concentration of 1–3 µg/µl. Purified protein was mixed with 11 kDa F-dextran (Sigma, St Louis, MO) to give a final F-dextran concentration of 1 mM and stored at 4°C until used. Chemically synthesized and HPLC-purified 12mer oligopeptides resembling KN1pep (KN1pepsynth and KN1-Npepsynth) and CMVpep2 and 3 (CMVpep2synth and CMVpep3synth) were purchased from United Biochemical Research, Inc. (UBR).
Phage display screens were carried out using the Ph.D.-12 Phage Display Peptide Library Kit (New England BioLabs, Beverly, MA). A tobacco plasmodesmal-enriched cell wall fraction (W2 fraction) was prepared as described previously (Kragler et al., 1998) and W2 fraction protein aliquots (10 µg) were then immobilized on precut nitrocellulose membranes (1 × 0.5 cm), placed in microcentrifuge tubes and allowed to dry. All of the following steps were carried out at 22°C. Membranes were blocked for 30 min in 1 ml of incubation buffer [40 mM HEPES pH 6.8, 10% (v/v) glycerol, 2 mM MgCl2, 1% (w/v) BSA] prior to a 30 min incubation in a mixture of phage library stock (10 µl aliquots of stock diluted into 1 ml of incubation buffer). Unbound phages were removed from the membrane using three 10 s washes with incubation buffer (1 ml). Elution of bound phages was achieved by addition of 20 µg/ml KN1, CMV-MP or E.coli lysate (as a control). After a 5 min incubation period, the resultant phage suspension was collected and stored at 4°C until used for amplification. Five rounds of panning and amplification were performed. Individual phages obtained from each treatment were then isolated and prepared for ssDNA sequencing. Phage particles were also precipitated, according to the manufacturer’s instructions, resuspended in protein buffer (1 µg/µl) and then stored at 4°C until used in microinjection and dot-blot assays.
Dot-blot assays
Plasmodesmal-enriched W2 cell fractions (100 ng/well) were transferred to nitrocellulose membrane (0.45 µm NitroBind, Micron Separations, Westborough, MA) prewetted with blotting buffer [40 mM HEPES pH 6.8, 10% (v/v) glycerol] using a dot-blot apparatus (Bio-Rad). Immobilized W2 fraction proteins were incubated in binding buffer [blotting buffer containing 5% (w/v) BSA and 2 mM MgCl2], which was removed after 30 min and replaced by a protein-gold particle preparation (200 ng/well); KN1-gold and CMV-MP-gold were prepared as described previously (Kragler et al., 1998). After 1 h incubation at 22°C the nitrocellulose membrane was washed (twice) with binding buffer (–BSA), and bound protein-gold complexes were fixed for 10 min with 1% (v/v) glutaraldehyde in binding buffer (–BSA). The nitrocellulose membrane was then washed (three times) with glass-distilled water, and incubated for 10 min with 50 mM EDTA pH 4.5. After a final rinse with glass-distilled water, silver enhancement was performed with LI Silver Stain solution (Nanoprobes, Stony Brook, NY) according to the manufacturer’s protocol, with the exception that these reactions were performed in the dark. Nitrocellulose membranes were immediately scanned for analysis with NIH Image 1.16 image processing software.
RNA isolation and labeling
Fluorescent KN1 RNA was transcriptionally labeled according to the manufacturer’s protocol (Maxiscript; Ambion, Austin, TX) using fluorescently labeled UTP-12-CF (Molecular Probes, Eugene, OR). Non-incorporated nucleotides were separated by repeated EtOH precipitation. The integrity, length (1.2 kb) and concentration of the RNA transcript were verified by agarose gel electrophoresis. Fluorescently labeled KN1–sense RNA-CF probes were resuspended at a final concentration of 2 µg/µl in protein buffer and stored at –20°C until used in microinjection experiments.
Microinjection procedures
Eight-week-old N.benthamiana plants were used for microinjection studies, with a minimum of five plants used per probe tested. Leaves (3–5 cm in length) were excised and prepared for microinjection experiments as described previously (Rojas et al., 1997). Cell-to-cell movement of fluorescently labeled probes was observed and recorded with either a Leitz Orthoplan epi-illumination fluorescence microscope, fitted with an analytical video imaging system (model VIM C1966-20; Hamamatsu Photonics, Bridgewater, NJ), or using a confocal laser scanning microscope (Leica model DM RXE TCS-4D). Spatial distribution of fluorescence within the mesophyll tissue was evaluated 5 min after the probe(s) was introduced into a target cell by pressure-mediated delivery.
Acknowledgments
Acknowledgements
We thank the members of our laboratory for insightful discussions throughout the course of this work. This work was supported by DOE Biosciences grant DE-FG03-94ER20134 and NSF grant IBN-9900539 (to W.J.L.). F.K. was supported by an Erwin Schrödinger Fellowship (J1249-GEN) provided by the Austrian FWF.
References
- Balachandran S., Xiang,Y., Schobert,C., Thompson,G.A. and Lucas,W.J. (1997) Phloem sap proteins of Cucurbita maxima and Ricinus communis have the capacity to traffic cell to cell through plasmodesmata. Proc. Natl Acad. Sci. USA, 94, 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berna A., Gafny,R., Wolf,W., Lucas,W.J., Holt,C.A. and Beachy,R.N. (1991) The TMV movement protein: Role of the C-terminal 73 amino acids in subcellular localization and function. Virology, 182, 682–689. [DOI] [PubMed] [Google Scholar]
- Bogerd H.P., Fridell,R.A., Benson,R.E., Hua,J. and Cullen,B.R. (1996) Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol., 16, 4207–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B. (1998) Intercellular protein trafficking through plasmodesmata. Plant Mol. Biol., 38, 279–310. [PubMed] [Google Scholar]
- Ding B., Itaya,A. and Woo,Y.M. (1999) Plasmodesmata and cell-to-cell communication in plants. Int. Rev. Cytol., 190, 251–316. [Google Scholar]
- Doorbar J. and Winter,G. (1994) Isolation of a peptide antagonist to the thrombin receptor using phage display. J. Mol. Biol., 244, 361–369. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Giesman-Cookmeyer,D., Ding,B., Lommel,S.A. and Lucas,W.J. (1993) Cell-to-cell trafficking of macromolecules through plasmodesmata potentiated by the red clover necrotic mosaic virus movement protein. Plant Cell, 5, 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafny R., Lapidot,M., Berna,A., Holt,C.A., Deom,C.M. and Beachy,R.N. (1992) Effects of terminal deletion mutations on function of the movement protein of tobacco mosaic virus. Virology, 187, 499–507. [DOI] [PubMed] [Google Scholar]
- Ghoshroy S., Lartey,R., Sheng,J.S. and Citovsky,V. (1997) Transport of proteins and nucleic acids through plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol., 48, 25–48. [DOI] [PubMed] [Google Scholar]
- Goodson R.J., Doyle,M.V., Kaufman,S.E. and Rosenberg,S. (1994) High-affinity urokinase receptor antagonists identified with bacteriophage peptide display. Proc. Natl Acad. Sci. USA, 91, 7129–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A., Truernit,E. and Sauer,N. (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell, 11, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwatari Y., Fujiwara,T., McFarland,K.C., Nemoto,K., Hayashi,H., Chino,M. and Lucas,W.J. (1998) Rice phloem thioredoxin h has the capacity to mediate its own cell-to-cell transport through plasmodesmata. Planta, 205, 12–22. [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit,B. and Hake,S. (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development, 120, 405–413. [Google Scholar]
- Kaplan I.B., Shintaku,M.H., Zhang,L., Marsh,L.E. and Palukaitis,P. (1995) Complementation of virus movement in transgenic tobacco expressing cucumber mosaic virus 3a gene. Virology, 209, 188–199. [DOI] [PubMed] [Google Scholar]
- Katz B.A. (1997) Structural and mechanistic determinants of affinity and specificity of ligands discovered or engineered by phage display. Annu. Rev. Biophys. Biomol. Struct., 26, 27–45. [DOI] [PubMed] [Google Scholar]
- Kragler F., Monzer,J., Shash,K., Xoconostle-Cázares,B. and Lucas,W.J. (1998) Cell-to-cell transport of proteins: Requirement for unfolding and characterization of binding to a putative plasmodesmal receptor. Plant J., 15, 367–381. [Google Scholar]
- Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lazarowitz S.G. and Beachy,R.N. (1999) Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell, 11, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-Y., Yoo,B.-C. and Lucas,W.J. (2000) Parallels between nuclear pore and plasmodesmal trafficking of information molecules. Planta, 210, 177–187. [DOI] [PubMed] [Google Scholar]
- Lough T.J., Shash,K., Hofstra,K.R., Xoconostle-Cázares,B., Beck,D.L., Balmori,E., Forster,R.L.S. and Lucas,W.J. (1998) Molecular dissection of the mechanism by which potexvirus triple gene block proteins mediate cell-to-cell transport of infectious RNA. Mol. Plant Microbe Interact., 11, 801–814. [Google Scholar]
- Lucas W.J. (1995) Plasmodesmata: Intercellular channels for macromolecular transport in plants. Curr. Opin. Cell Biol., 7, 673–680. [DOI] [PubMed] [Google Scholar]
- Lucas W.J. (1999) Plasmodesmata and the cell-to-cell transport of proteins and nucleoprotein complexes. J. Exp. Bot., 50, 979–987. [Google Scholar]
- Lucas W.J., Bouché-Pillon,S., Jackson,D.P., Nguyen,L., Baker,L., Ding,B. and Hake,S. (1995) Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science, 270, 1980–1983. [DOI] [PubMed] [Google Scholar]
- McLean B.G., Hempel,F.D. and Zambryski,P.C. (1997) Plant intercellular communication via plasmodesmata. Plant Cell, 9, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher U. (1990) Similarities between putative transport proteins of plant viruses. J. Gen. Virol., 71, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Mezitt L.A. and Lucas,W.J. (1996) Plasmodesmal cell-to-cell transport of proteins and nucleic acids. Plant Mol. Biol., 32, 251–273. [DOI] [PubMed] [Google Scholar]
- Noueiry A.O., Lucas,W.J. and Gilbertson,R.L. (1994) Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell, 76, 925–932. [DOI] [PubMed] [Google Scholar]
- Oparka K.J., Roberts,A.G., Boevink,P., Santa Cruz,S., Roberts,I.M., Kotlizky,G., Sauer,N. and Epel,B. (1999) Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell, 97, 743–754. [DOI] [PubMed] [Google Scholar]
- Peters E.A., Schatz,P.J., Johnson,S.S. and Dower,W.J. (1994) Membrane insertion defects caused by positive charges in the early mature region of protein pIII of filamentous phage fd can be corrected by prlA suppressors. J. Bacteriol., 176, 4296–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser L., Sanchez-Baracaldo,P. and Hake,S. (2000) Knots in the family tree: evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol., 42, 151–166. [PubMed] [Google Scholar]
- Robards A.W. and Lucas,W.J. (1990) Plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol., 41, 369–419. [Google Scholar]
- Rojas M.R., Zerbini,F.M., Allison,R.F., Gilbertson,R.L. and Lucas,W.J. (1997) Capsid protein and helper component-proteinase function as potyvirus plasmodesmal movement proteins. Virology, 237, 283–295. [DOI] [PubMed] [Google Scholar]
- Schatz G. (1998) Protein transport—the doors to organelles. Nature, 395, 439–440. [DOI] [PubMed] [Google Scholar]
- Subramani S. (1996) Protein translocation into peroxisomes. J. Biol. Chem., 271, 32483–32486. [DOI] [PubMed] [Google Scholar]
- Szardenings M., Tornroth,S., Mutulis,F., Muceniece,R., Keinanen,K., Kuusinen,A. and Wikberg,J.E. (1997) Phage display selection on whole cells yields a peptide specific for melanocortin receptor 1. J. Biol. Chem., 272, 27943–27948. [DOI] [PubMed] [Google Scholar]
- Terry B.R. and Robards,A.W. (1987) Hydrodynamic radius alone governs the mobility of molecules through the plasmodesmata. Planta, 171, 145–157. [DOI] [PubMed] [Google Scholar]
- Waigmann E. and Zambryski,P. (1995) Tobacco mosaic virus movement protein-mediated protein transport between trichome cells. Plant Cell, 7, 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E., Lucas,W.J., Citovsky,V. and Zambryski,P. (1994) Direct functional assay for tobacco mosaic virus cell-to-cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc. Natl Acad. Sci. USA, 91, 1433–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Deom,C.M., Beachy,R.N. and Lucas,W.J. (1989) Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science, 246, 377–379. [DOI] [PubMed] [Google Scholar]