Abstract
NaeI is transformed from DNA endonuclease to DNA topoisomerase and recombinase by a single amino acid substitution. The crystal structure of NaeI was solved at 2.3 Å resolution and shows that NaeI is a dimeric molecule with two domains per monomer. Each domain contains one potential DNA recognition motif corresponding to either endonuclease or topoisomerase activity. The N-terminal domain core folds like the other type II restriction endonucleases as well as λ-exonuclease and the DNA repair enzymes MutH and Vsr, implying a common evolutionary origin and catalytic mechanism. The C-terminal domain contains a catabolite activator protein (CAP) motif present in many DNA-binding proteins, including the type IA and type II topoisomerases. Thus, the NaeI structure implies that DNA processing enzymes evolved from a few common ancestors. NaeI may be an evolutionary bridge between endonuclease and DNA processing enzymes.
Keywords: CAP/crystal structure/DNA topoisomerase/helix–turn–helix/NaeI restriction endonuclease
Introduction
DNA endonucleases, topoisomerases, recombinases and ligases are ubiquitous enzymes essential for genetic processes such as replication, transcription, recombination and repair of DNA. Restriction endonucleases in prokaryotes protect the host against invading genomes, showing a great ability to recognize and cleave short specific DNA sequences hidden within the large background of DNAs. Over 3000 restriction endonucleases are known, representing >200 different sequence specificities (reviewed by Roberts and Macelis, 2000). Restriction enzymes show little sequence homology either with each other or with other protein families, although possible homology was found between EcoRII and the integrase family (Topal and Conrad, 1993). There are three types of restriction endonucleases (reviewed by Wilson, 1991). Type I enzymes are multimeric proteins that contain both endonuclease and methylase activities, recognize specific sequences and cleave at distant random sites outside the recognition sequence. ATP and S-adenosyl-methionine are required cofactors. Type II enzymes, major contributors to the biotechnology revolution, are dimeric proteins that recognize and cleave within specific, usually palindromic, DNA sequences 4–8 bp in length. Type III enzymes are complexes of restriction and methylation subunits, require ATP binding but not hydrolysis, and cleave ∼25 bp away from the recognition sequence.
NaeI was isolated from a strain of the actinomycete Nocardia aerocolonigenes (ATCC 23870) cultured from soil in Japan (Shinobu and Kawato, 1960; Labeda, 1986). NaeI is a type IIe endonuclease (Yang et al., 1994) with unique properties. It is allosteric (Conrad and Topal, 1989; Yang and Topal, 1992), binding two GCC↑CGG recognition sequences to cleave DNA into blunt-ended products. NaeI is a prototype of the growing number of type IIe endonucleases (Krüger et al., 1988; Conrad and Topal, 1989; Oller et al., 1991; Reuter et al., 1993) and is the first restriction endonuclease found to form a covalent intermediate with its DNA substrate (Jo and Topal, 1995). A 10 amino acid sequence in the N-terminal domain of NaeI is similar to the active site of DNA ligase I, except for Leu43 in NaeI in place of the lysine essential for ligase function. Remarkably, substitution L43K converts NaeI from endonuclease to topoisomerase and recombinase (Jo and Topal, 1995). Thus, NaeI appears to be a missing link that relates these DNA-binding proteins. We report here the structure of apoNaeI at 2.3 Å resolution, the first structure of a type IIe endonuclease.
Results and discussion
Two different DNA-recognition structural motifs
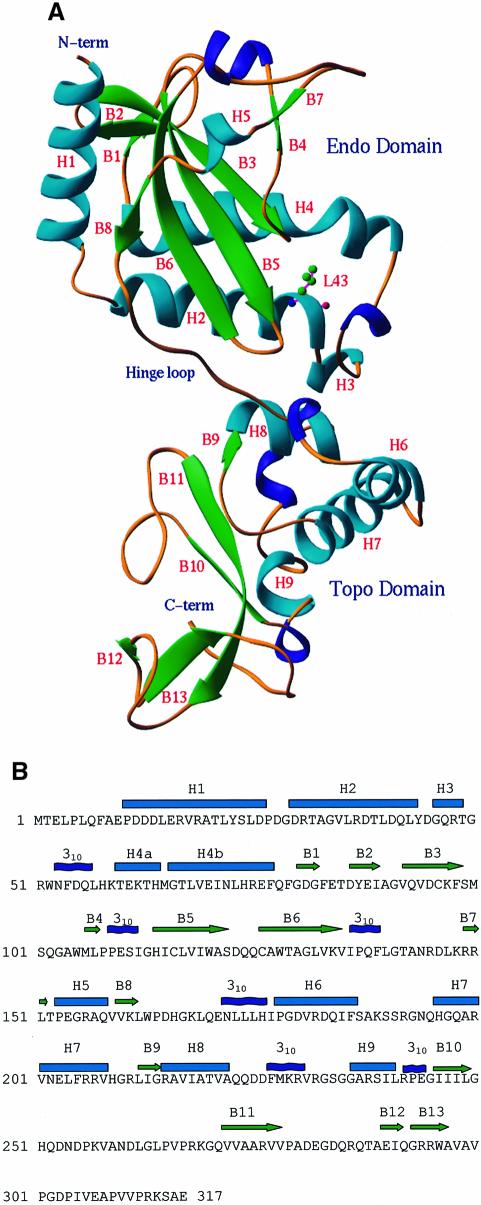
The structure of the NaeI monomer consists of nine α-helices, six 310 helices and 13 β-strands, which aggregate into two domains (Figure 1A). The N-terminal domain is composed of residues 10–162 (the first nine residues were not observed in the structure) and has approximate dimensions 27 × 32 × 38 Å3. It contains a core structure of a six-stranded β-sheet flanked with five helices on one side and two strands and three helices on another side. Random mutagenesis of NaeI shows that most if not all amino acids involved in DNA cleavage reside in the N-terminal domain (Holtz and Topal, 1994), suggesting that the N-terminal domain is the active site of the endonuclease. We tentatively name the N-terminal domain the ‘Endonuclease’ (Endo) domain for its topological similarity to other type II restriction endonucleases.
Fig. 1. Structure of the NaeI monomer. (A) Ribbon diagram: α-helices in cyan, 310 helices in blue and β-strands in green. The N-terminal (Endo) domain has topology similar to other restriction endonucleases. The C-terminal (Topo) domain contains a CAP or helix–turn–helix motif (H7, H8, 310 helix before H9, B10, B11) for DNA binding. The hinge loop is around Gln170. L43 is shown in ball-and-stick representation. (B) Sequence and secondary structure of NaeI. Cyan cylinders and green arrows represent α-helices and β-strands, respectively. The helices H4a and H4b are bent at Met65.
The C-terminal domain of NaeI (residues 172–311) consists of a two-layer β-sandwich adjacent to four helices (Figure 1), and has approximate dimensions 28 × 31 × 42 Å3. The domain contains the DNA-binding motif of catabolite activator protein (CAP). CAP is a helix–turn–helix (HTH) motif observed in many DNA processing proteins (reviewed by Harrison, 1991; Pabo and Sauer, 1992; Nelson, 1995), including the type IA and type II DNA topoisomerases (reviewed by Berger et al., 1998). Finding this structural link between NaeI and topoisomerases may be especially important because the L43K mutation gives NaeI topoisomerase activity. We tentatively name the C-terminal domain the ‘Topoisomerase’ (Topo) domain.
The Endo domain of NaeI was well ordered, whereas the Topo domain showed several disordered loops and had relatively higher B-factors. Topo domain residues 191–194, 254–258 and 312–317 were not observable in the structure, compared with only missing Endo domain residues 1–9. The average B-factor was 61.6 Å2 for all atoms of the Topo domain, in comparison with 41.2 Å2 for those of the Endo domain. Thus, the Topo domain appears to be loosely packed and to possess significant conformational flexibility. The Endo and Topo domains are linked by a relatively extended hinge loop including Tyr163–Glu171. The two domains pack against one another via van der Waals interactions as well as a few hydrogen bonds.
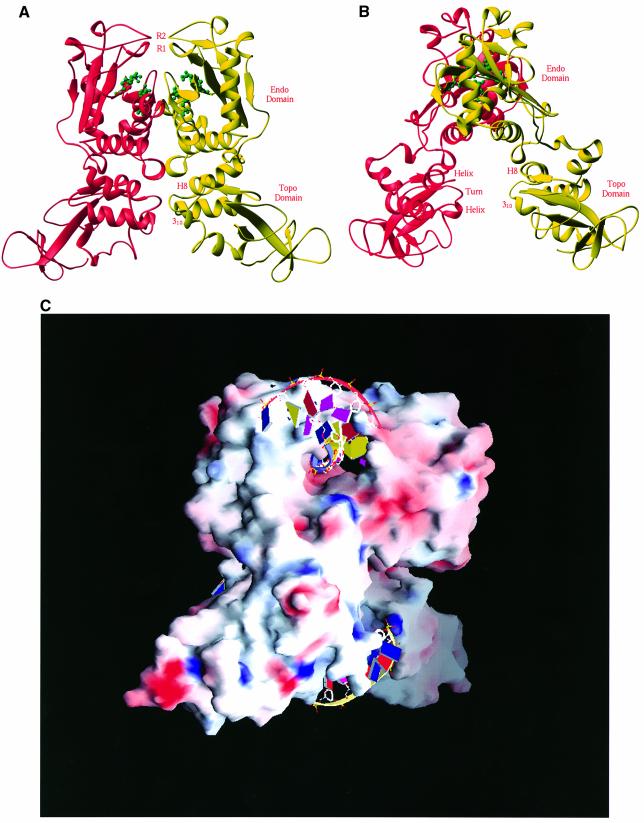
Two monomeric molecules of NaeI are tightly associated into a dimer via interfacial interactions between helices H4 of each molecule, and are related by a local molecular 2-fold axis (Figure 2). Dimerization of the Endo domains forms a cleft that has rough dimensions of 15 Å deep by 22 Å wide and that is covered by two fragments around B4 and B7 (Figure 2). This cleft contains the active site residues of NaeI and is the endonuclease site of NaeI, based on its similarity to the active sites of other type II restriction endonucleases. The Topo domains of the NaeI dimer show much less interdomain interaction and form a cleft with ∼13 Å between the HTH motifs (Figure 2B). The 310 helix before H9 is likely to be the recognition helix for DNA binding in a manner similar to the CAP motifs of the topoisomerases. This suggests that the Topo domain is responsible for the topoisomerase activity of NaeI L43K (see discussion below). The assignment of the Endo domain to substrate binding implies that the Topo domain binds activator DNA and is responsible for initiating the conformational changes that enable DNA cleavage (Conrad and Topal, 1989; Yang and Topal, 1992).
Fig. 2. Structure of the NaeI dimer. (A) Two monomers in red and yellow ribbons are related by a molecular 2-fold axis located vertically. The tentative active site residues of Glu70, Asp86, Asp95 and Lys97 for the endonuclease activity are shown in green ball-and-stick representation. Two putative recognition loops are located at the very top of the picture, and are marked R1 (residues 102–109, β-strand B4) and R2 (residues 141–149, a β-like turn). (B) A view of the NaeI dimer with a 90° rotation about the vertical axis from (A). The CAP DNA-binding motif in the Topo domain of NaeI, labeled helix–turn–helix and H8 and 310 (before H9), is common for many of the DNA-binding proteins such as topoisomerases and transcription factors. This site is presumably a second site of DNA binding and responsible for the topoisomerase activity of the NaeI L43K mutant. (C) Surface presentation of dimeric NaeI: red for negatively charged residues and blue for positively charged residues. Two DNA fragments are modeled into the endonuclease (top) and topoisomerase (bottom) sites on the basis of the superposition of NaeI over the PvuII–DNA and CAP–DNA structures, respectively.
Conformational asymmetry of the NaeI dimer
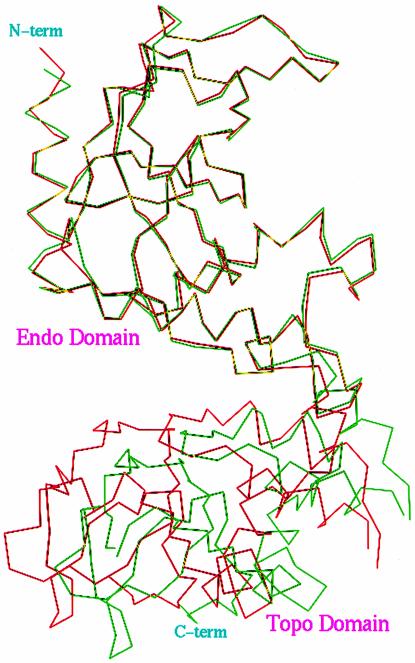
Careful inspection of the NaeI dimer revealed that the relative orientations of the 2-fold axes for the Endo and Topo domains in the NaeI dimer are slightly different, with ∼3° rotational offset and >10 Å translation. Superposition of the backbone atoms of the NaeI homodimer revealed root mean square (r.m.s.) displacements of 0.63 and 1.04 Å for the Endo and Topo domains, respectively, and 2.02 Å for the entire homodimer. When the transformation matrix that best superimposed the Endo domains was applied to the Topo domains, an overall r.m.s. separation of 5.04 Å was observed for 504 backbone atoms of the Topo domain (Figure 3). This indicates that the quaternary structure or the domain orientation in each monomer of NaeI is significantly different.
Fig. 3. Cα trace for the superposition of two monomers (red and green) in the NaeI dimer. The transformational matrix of the Endo domain was used for the superposition of the entire molecule. The superposition showed ∼5 Å displacement for the backbone atoms of the Topo domain, indicating that the Endo and Topo domains have significantly different orientation of their 2-fold axes.
Since the Endo domain compares well to dimers of other restriction endonucleases and the Topo domain is loosely packed, the conformational asymmetry is most likely to be dependent on the orientation of the Topo domain. The asymmetry does not appear to be due to crystal packing because the Topo domains are not heavily involved in the formation of the crystal lattice, thus implying that the asymmetry represents a native conformational state of the enzyme. The biological implications of this asymmetry are unknown. Comparison of the two hinge loops in the dimer failed to detect a dramatic conformational change associated with a single residue. Instead, the conformational differences appear to be distributed over much of the Tyr163–Glu171 loop. This reinforces the notion that these residues serve as a hinge region to alter molecular conformation and enable communication between the domains during catalysis.
Evolutionary origin of nucleases
Restriction endonucleases generally show very little sequence homology with each other or with other endo- or exonucleases. Reported structures for the type II restriction endonucleases, however, possess a common core topological motif (CCM; Venclovas et al., 1994; Newman et al., 1995). Structural superposition using the DALI (Holm and Sander, 1993) and CCP4 programs detected similarities of NaeI with eight restriction endonucleases, DNA repair endonucleases Vsr (very short patch repair) and MutH (mismatch repair), and the 5′–3′ λ-exonuclease (recombination and repair). The superposition between these enzymes showed r.m.s. deviations of ∼2 Å for the backbone atoms of the 21–71 compared residues, implying structural conservation of the molecular cores of the nucleases.
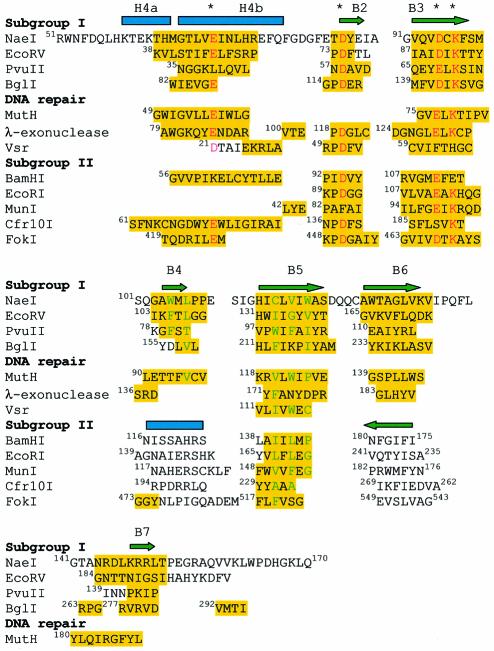
Structure-based sequence alignment revealed absolute conservation of three β-strands in all 12 compared structures: B2, B3 and B5 in NaeI (Figure 4). Two of these β-strands, equivalent to B2 and B3 in NaeI, bind divalent metals and DNA substrate (McClarin et al., 1986; Kim et al., 1990; Winkler et al., 1993; Cheng et al., 1994; Kostrewa and Winkler, 1995; Newman et al., 1995) and thus their conservation is essential for nuclease activity. NaeI β-strand B5, or the equivalent in the other endonucleases, is apparently not involved in metal binding and DNA cleavage (Figure 4), but may be critical for formation of the β-sheet and the hydrophobic core of the nuclease molecules. For example, NaeI Cys116 and Val118 in B5 and Trp105 and Leu107 in B4 form the hydrophobic core of the NaeI Endo domain. This hydrophobic core appears necessary to place loop 102–109 correctly for DNA recognition by NaeI (see discussion below). Thus, we argue, based on their apparently essential structural and catalytic roles for endonuclease activity, that the three conserved β-strands are the common origin of divergent evolution for the nucleases. This differs from the prevailing view that the endonuclease CCM consists of a five-stranded β-sheet flanked by two α-helices (Venclovas et al., 1994; Kovall and Matthews, 1999).
Fig. 4. Structure-based sequence alignment of the NaeI Endo domain with partial sequences of eight restriction endonucleases, two DNA repair endonucleases and λ-exonuclease. Amino acids highlighted in yellow are used in the calculation of the r.m.s. deviation of the superposition. Three negatively charged residues for divalent metal binding and the catalytic lysine are marked with asterisks and colored red. Hydrophobic residues of the molecule core are colored green. The alignment suggests that the restriction endonucleases are divided into subgroup I (NaeI, EcoRV, PvuII and BglI) and subgroup II (BamHI, EcoRI, MunI, Cfr10I and FokI). Subgroup II shows a β-strand running in an opposite direction to B6 in NaeI, and also a helix in place of β-strand B4 in NaeI. B4 and B7 in NaeI are, respectively, contained in recognition loops R1 and R2 (Figure 2A).
Also highly conserved is α-helix H4 in NaeI (Figure 4). All of the compared nucleases except for EcoRI and MunI have an equivalent helix, although longer or shorter, occupying a similar spatial location. H4, however, differs in function among the different enzymes. H4 in NaeI (residues Lys59–Phe77) mediates dimerization (Figures 1 and 2). The corresponding helix interacts only with DNA substrate in BamHI, and plays a dual role of dimerization and DNA binding in EcoRV, PvuII and BglI. Thus, helix H4 is apparently not an evolutionarily conserved essential element, but instead a consequence of divergent evolution among nucleases.
Conservation and versatility of catalytic mechanisms
Structure-based sequence alignment identified four conserved NaeI residues: Glu70, Asp86, Asp95 and Lys97 (Figure 4). Residues equivalent to Asp86 and Asp95 of NaeI have been identified as the divalent metal-binding site in the enzyme–DNA–metal complexes of BamHI (Newman et al., 1995; Viadiu and Aggarwal, 1998), EcoRI (McClarin et al., 1986; Kim et al., 1990), EcoRV (Horton et al., 1998; Martin et al., 1999) and BglI (Newman et al., 1998). The conserved lysine appears to stabilize the doubly charged pentavalent transition state (Kostrewa and Winkler, 1995) and to orient the attacking water molecule (Horton et al., 1998). Alignment of the nuclease catalytic motifs supports a common catalytic mechanism among these enzymes, in which the divalent metal stabilizes the DNA binding and possibly activates a water to attack the scissile phosphate directly, resulting in a pentavalent transition state stabilized by the conserved lysine (reviewed by Kovall and Matthews, 1999). This general scheme of catalysis has also been proposed for other DNA processing enzymes. For example, T7 polymerase uses two magnesium ions for binding of the DNA and an arginine for stabilization of the transition state (Doublie et al., 1998).
The structure-based sequence alignment also showed that DNA repair enzymes MutH (Ban and Yang, 1998), Vsr (Tsutakawa et al., 1999) and λ-exonuclease (Kovall and Matthews, 1997, 1998) have Glu56/Glu77/Lys79, Asp21/Asp51/His64 and Glu85/Asp119/Glu129/Lys131, respectively, at or near their metal-binding sites (Figure 4). This implies that these DNA processing enzymes share a similar catalytic mechanism with the type II restriction endonucleases. In that light, it is interesting that a Vsr homolog is part of the NaeI operon (Taron et al., 1995), strengthening the link between these two activities.
On the other hand, the structural superposition also shows divergence in catalytic residues among the endonucleases. First, a negatively charged residue Glu/Asp at the topological position Glu70 in NaeI (Figure 4), a residue generally considered for metal binding (Kovall and Matthews, 1999), is substituted by Lys61 in BamHI and by Leu39 in PvuII. Secondly, position Asp95 in NaeI is occupied by Ser188 in Cfr10I (Bozic et al., 1996). These substitutions suggest that different protein residues are involved in the metal binding in these enzymes. Thirdly, Lys97 in NaeI, assumed to stabilize the catalytic intermediate in EcoRV (Horton et al., 1998), is substituted by Glu113 in BamHI (Figure 4), suggesting the versatility of the detailed reaction pathway (see the review by Kovall and Matthews, 1999).
Two endonuclease subgroups
The structures of type II endonucleases have been divided into two subgroups based on whether the respective endonucleases cleave their cognate recognition sequences to give 5′ overhanging versus blunt-ended products (Pingoud and Jeltsch, 1997; Kovall and Matthews, 1999). Our structural alignment, however, suggests that endonucleases are better categorized by the orientation of β-strand B6 in NaeI and by the occurrence of β-strand versus α-helix at position B4 in NaeI (Figure 4). The endonucleases in subgroup I (NaeI, BglI, EcoRV and PvuII) contain two β-strands that correspond to and have the same chain polarity as B6 and B4 in NarI. On the other hand, the endonucleases in subgroup II (EcoRI, BamHI, Cfr10I, MunI and FokI) show β-strands with an opposite chain direction to β-strand B6 of subgroup I, and α-helices in place of β-strand B4 in NaeI (Figure 4). By this classification, the secondary structure elements show maximum comparability within each subgroup but minimal comparability across the subgroups.
Superposition of a monomer of NaeI over that of either EcoRV or PvuII automatically superimposed the second monomer. Similar superposition of NaeI with any other endonucleases of Figure 4, however, placed the second monomer at very different locations. This suggests that NaeI, EcoRV and PvuII have a similar dimerization scheme, probably needed for the blunt cleavage of their recognition sequence.
Two different patterns of DNA recognition
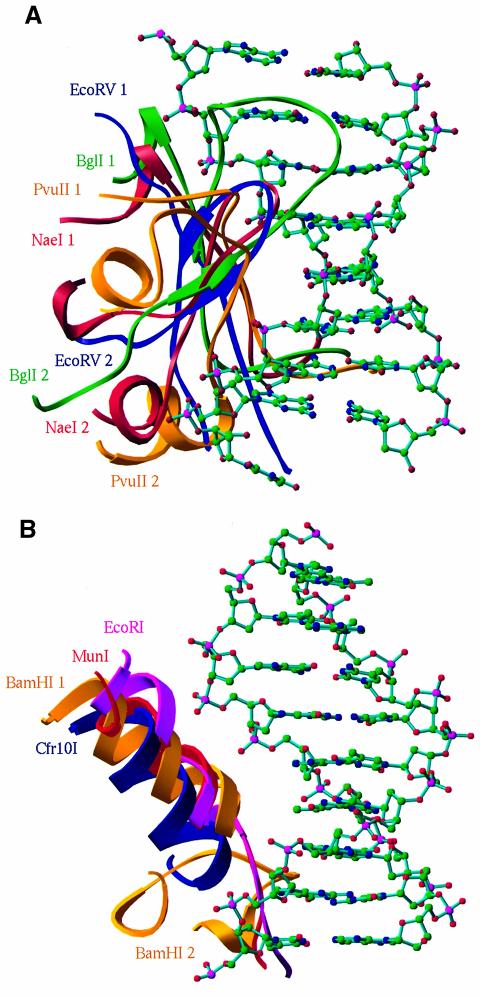
DNA recognition elements have been reported, but not systematically compared for EcoRI (McClarin et al., 1986; Kim et al., 1990), BamHI (Newman et al., 1995), MunI (Deibert et al., 1999), EcoRV (Winkler et al., 1993) and PvuII (Cheng et al., 1994). Our structural superposition demonstrates that these recognition elements, although they have different amino acid components and recognize different DNA sequences, occupy the same location to interact with the major groove of the cognate DNAs (Figure 5). The recognition can be grouped into two patterns. The members in subgroup I, including NaeI, apparently use a β-strand and a β-like turn for DNA recognition (R1 and R2 in Figures 2A, 4 and 5A). We tentatively call it β-strand recognition. In contrast, those in subgroup II use an α-helix and a loop (Figures 4 and 5B). We tentatively call the latter α-helix recognition, to differentiate between the two types of DNA recognition by endonucleases. We predict that these two patterns of recognition may apply to most, if not all, type II restriction endonucleases.
Fig. 5. Two patterns of DNA recognition of the endonucleases. (A) The β-strand recognition for subgroup I endonucleases consists of two elements: a β-strand (NaeI 1 in this figure and R1 in Figure 2A) and a β-like turn (NaeI 2 in this figure and R2 in Figure 2A). The color codes are red for NaeI (100–113 and 135–158), gold for PvuII (75–88 and 129–152), green for BglI (149–162 and 256–279) and blue for EcoRV (101–113 and 175–195). The 8 bp double helix DNA is adopted from the PvuII–DNA structure (Cheng et al., 1994). (B) The helix recognition for subgroup II endonucleases of BamHI (gold; BamHI 1 for 113–126 and BamHI 2 for 145–165), Cfr10I (blue; 192–206), EcoRI (magenta; 137–152) and MunI (red; 113–128). The double helix DNA is adopted from the BamHI–DNA structure (Newman et al., 1995).
It is interesting to note that the DNA mismatch repair enzyme MutH contains two fragments, Leu90–Val97 and Tyr180–Leu188, corresponding to the putative recognition sequences Gln102–Pro109 and Gly141–Arg149 of NaeI (Figure 4). This implies that MutH probably has a β-strand recognition mechanism in a manner similar to subgroup I, although monomeric MutH cleaves only one strand of the DNA duplex in comparison to the cleavage of both DNA strands by dimeric type II endonucleases.
CAP motif of NaeI Topo domain
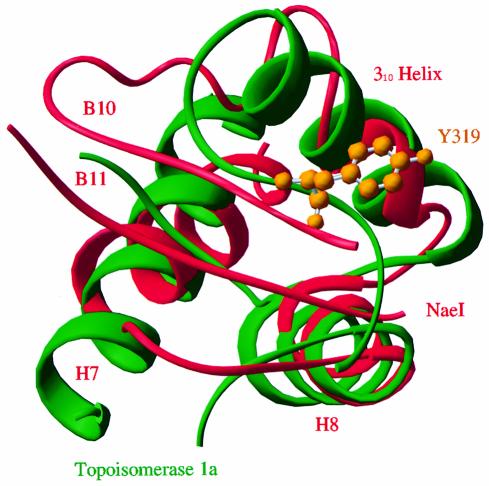
The DNA-binding motif of the Escherichia coli CAP, also known as an HTH motif, is found in many DNA-binding proteins (reviewed by Harrison, 1991; Pabo and Sauer, 1992; Nelson, 1995; Berger et al., 1998). In the Topo domain of NaeI, the α-helices H7 and H8, a 310 helix before H9 and two β-strands B10 and B11 form a structural scaffold resembling CAP. A structural comparison program, DALI (Holm and Sander, 1993), revealed that the CAP in NaeI is superimposable with DNA-binding domains of many DNA processing proteins, as shown in a partial list of topoisomerases Ia and II [Protein Data Bank (PDB) entry 1rva and 1bgw], catabolite gene activator (2cgp), transcription regulators of E2F-DP (1cf7), MotA (1bja), OMPR (1opc), Smtb (1smt) and LexA (1lea), RNA adenosine deaminase Zα (1qbj), mating-type protein MATA1 (1akh), nitrate/nitrite regulator NarL (1a04), chromosomal protein histone H5 (1hst) and restriction endonuclease FokI (2fok). DNA binding by each protein commonly involves the third helix of CAP, or the 310 helix before H9 in NaeI (Figure 6). Structure-based sequence alignment of the CAP motifs of these proteins showed no conservation of sequence (data not shown). This is not surprising considering the variety of sequences for DNA recognition. The structural homologies imply that the CAP motif of NaeI is a DNA-binding site. Indeed, the NaeI Topo domain, cloned and isolated from E.coli, specifically binds cognate DNA recognition sequence with an ∼10-fold lower affinity than wild-type NaeI (Colandene and Topal, 1998).
Fig. 6. Superposition of the CAP motifs of NaeI (red) and topoisomerase IA (green). The active site Tyr319 of topoisomerase IA, which forms a covalent intermediate with DNA substrate, is shown as gold balls. The 310 helix before H9 in NaeI corresponds to the helix that binds the major groove of DNA in CAP DNA-binding proteins.
Putative topoisomerase mechanism of NaeI
NaeI protein contains the sequence 39TLDQLYDGQR48, which is similar to a fragment at the active site of DNA ligase I except for leucine in place of the ligase essential lysine (Jo and Topal, 1995). The single mutation L43K gives NaeI DNA topoisomerase activity (Jo and Topal, 1995, 1998). Structural comparison showed that NaeI and topoisomerases IA and II possess a common CAP motif (Figure 6). The CAP domains of topoisomerases IA and II are implicated in DNA binding and contain an active site tyrosine that forms covalent phosphotyrosyl intermediates for cleavage of DNA (Lima et al., 1994; Berger et al., 1996, 1998; Keck and Berger, 1999). Thus, the CAP motif of NaeI might explain the topoisomerase activity of the NaeI L43K mutant. The catalytic residues Tyr319 of topoisomerase IA and Tyr783 of topoisomerase II, however, correspond to Leu249 and Gly270, respectively, in NaeI. In fact, there is no tyrosine located in the Topo domain of NaeI, suggesting a different mechanism for the topoisomerase activity of the NaeI L43K mutant.
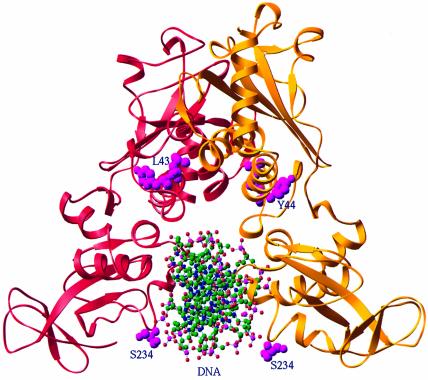
In the NaeI dimer, the two recognition helices of the CAP motif are separated by ∼13 Å (Figure 2B). Modeling the CAP–DNA structure (Schultz et al., 1991) onto the NaeI Topo domain showed that both 310 helices simultaneously interact with the same DNA fragment, and NaeI serine residues, instead of tyrosine, reside near the DNA-binding site (Figure 7), suggesting potential formation of a phosphoserinyl intermediate. The suggestion is consistent with the use of serine in place of tyrosine by the transposon γδ resolvase for covalent bond formation with DNA (Newman and Grindley, 1984). The best serine candidate for covalent interaction with the modeled DNA is Ser234, its side chain hydroxyl group sitting ∼5 Å away from the phosphodiester scissile bond of the DNA NaeI recognition sequence (Figure 7).
Fig. 7. Modeling of a DNA fragment onto the Topo domain of NaeI. The NaeI dimer is shown in red and gold ribbons, whereas DNA in the ball-and-stick model is adopted from the CAP–DNA structure. Ser234 and Tyr44 (magenta balls) are two potential candidates to form a covalent phosphoester intermediate for the topoisomerase activity of the L43K mutant. Leu43 is also shown as magenta balls.
On the other hand, two alternative mechanisms for the L43K topoisomerase activity are also possible. First, Leu43, as part of helix H2, lies at a position central to the hydrophobic core of the NaeI dimer and too far from DNA binding to be involved in the topoisomerase activity in the present structural conformation (Figure 7). The L43K substitution, however, may deform the NaeI dimer because of the intolerance of the hydrophobic core for the positively charged lysine. Thus, a dramatic conformational change may be induced by the L43K mutation, bringing Tyr44 to a position close to the CAP region to serve as the catalytic residue for topoisomerase activity (Figure 7). This is consistent with the overall picture that conformational change is an essential step for topoisomerase activity (Keck and Berger, 1999). Alternatively, the CAP motif could serve as a recognition site to anchor the DNA substrate while the topoisomerase activity takes place at the endonuclease site of the Endo domain. In this consideration, Tyr87 located near the metal-binding site might serve as the nucleophile to ligate DNA.
Concluding remarks
The crystal structure of NaeI at 2.3 Å resolution shows two domains, the Endo and Topo domains, which contain two structural motifs resembling the active site of restriction endonuclease and CAP DNA recognition. Structure-based sequence alignment demonstrated that the Endo domain of NaeI possesses a conserved core motif and catalytic residues in common with the other restriction endonucleases and with repair nucleases MutH, Vsr and λ-exonuclease. This suggests a common evolutionary origin of these nucleases. The Topo domain showed a CAP structure in common with other DNA-binding proteins such as topoisomerases IA and II, implying its role in the topoisomerase activity of the NaeI L43K mutant. The ability of NaeI to bridge the endonuclease and topoisomerase families suggests that NaeI is an evolutionary precursor to the type II restriction endonucleases, which have lost the Topo domain in return for embellishment of the Endo domain.
Materials and methods
Protein purification
Wild-type NaeI was purified as described previously (Colandene and Topal, 1998). Selenomethionyl-substituted NaeI was prepared following the method outlined by Doublie (1997). Briefly, E.coli strain CAA1 harboring plasmid pmalc2:NaeI (Colandene and Topal, 1998) was grown in LB culture medium at 37°C overnight. Cells were pelleted by centrifugation and resuspended in modified M9 minimal medium that was pre-warmed to 37°C. The modified medium was supplemented with the following amino acids (per liter): 100 mg Lys, 100 mg Thr, 100 mg Phe, 50 mg Leu, 50 mg Ile, 50 mg Val and 50 mg SeMet. Cells were grown at 37°C to an A595 of 0.5–0.7, and isopropyl-β-d-thiogalactopyranoside (IPTG) added to 0.2 mM. The temperature was adjusted to 20°C and the cells were grown for an additional 24–30 h. Purification of selenomethionyl NaeI from cell paste is the same as for wild-type NaeI (Colandene and Topal, 1998).
Crystallization
ApoNaeI and selenomethionyl NaeI were crystallized using microdialysis against a buffer of 20 mM malaic acid, 20 mM NaCl, 5 mM CaCl2, 1 mM β-mercaptoethanol, 2% PEG3350 and 1.5% ethanol at 4°C and pH 6.1. The crystals have the space group P21 with a = 99.4, b = 55.9, c = 59.0 Å and β = 95.7°, and contain two molecules of NaeI in the crystallographic asymmetric unit. The crystals have ∼0.6 mm in each dimension and almost perfect shape, but diffract only to moderate resolution.
NaeI endonuclease was crystallized after considerable difficulty. The crystals showed a relatively large mosaicity of ∼1, which suggests significant conformational flexibility of the NaeI molecule. Significant flexibility was also implied by the results of isoelectrofocusing. Multiple isoelectrofocusing bands were observed to cover ∼2 pH units for a single molecular weight of NaeI (a single band in SDS–PAGE). Precise control of pH, crystal growth conditions and cryo-cooling protocols gave crystals that diffracted to a resolution of ∼2.3 Å and enabled solution of the three-dimensional structure of the NaeI apomer.
Data collection and structure determination
For data collection, the crystals were soaked for 1 day in stabilization buffer, which contains the same components as the crystallization buffer but 15% PEG3350, and then transferred to 30% PEG3350 buffer for 2–6 h. The cryosolvent of 30% PEG3350 plus 5% PEG400 was used to freeze the crystals in liquid nitrogen. The best diffraction data for the native and selenomethionyl NaeI were collected at Brookhaven using synchrotron beam line X12C (Table I). All data were processed with program HKL (Otwinowski and Minor, 1997). The structure of NaeI was determined by multiwavelength anomalous diffraction (MAD) using selenomethionine as the anomalous scatterer. The automatic phasing program SOLVE 1.17 (Terwilliger and Berendzen, 1999) yielded an overall figure of merit of 0.52 for 20 893 reflections at 2.5 Å resolution and a partially traceable electron density map. The MAD phases were further improved by solvent flattening, histogram and local 2-fold symmetry averaging, as implemented in the CCP4 program package. The structure was modeled using the program FRODO and was refined by CNS (Brünger et al., 1998).
Table I. Crystallographic data statistics for native NaeI and the selenomethionyl mutant.
| Data | Wavelength (Å) | Resolution (Å) | Reflections |
Rsym | Completeness (%) | <I/σ> | |
|---|---|---|---|---|---|---|---|
| Total | Unique | ||||||
| Native | 0.97887 | 2.3 | 68 874 | 25 755 | 0.055 | 88.1 | 16.3 |
| SeNae(L1) | 0.97873 | 2.5 | 102 240 | 40 250 | 0.061 | 88.7 | 11.6 |
| SeNae(L2) | 0.97884 | 2.5 | 102 104 | 40 259 | 0.061 | 88.7 | 11.4 |
| SeNae(L3) | 0.95641 | 2.5 | 105 843 | 40 991 | 0.060 | 90.3 | 11.3 |
| Refinement | |||||||
| Space group | P21 | ||||||
| Unit cell | a = 99.4, b = 55.9 c = 59.0 Å and β = 95.7° | ||||||
| Resolution | 50–2.3 Å | ||||||
| Reflections | 25 672 (87.9%) | ||||||
| R-factor | 0.238 | ||||||
| R-free | 0.287 (8.7%) | ||||||
| Root mean square errors | |||||||
| bond length | 0.006 Å | ||||||
| angle | 1.29° | ||||||
| Protein residues | 10–190, 195–253, 259–311 | ||||||
| Water molecules | 91 | ||||||
| Average B-factors (Å2) | |||||||
| molecular A | 56.8 | ||||||
| molecular B | 42.8 | ||||||
| water | 33.1 | ||||||
Automatic MAD phasing by SOLVE resulted in a figure of merit of 0.52 at 2.5 Å resolution.
The atomic coordinates of NaeI have been deposited into the PDB with entry code 1ev7.
Acknowledgments
Acknowledgements
We would like to thank Dr R.Huber for the coordinates of MunI and Dr M.Newman for the coordinates of BglI. We thank Dr Sweet for help with diffraction data collection at beamline X12C at the National Synchrotron Light Source. This research is partially supported by NIH grant GM52123.
References
- Ban C. and Yang,W. (1998) Structural basis for MutH activation in E.coli mismatch repair and relationship of MutH to restriction endonucleases. EMBO J., 17, 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger J.M., Gamblin,S.J., Harrison,S.C. and Wang,J.C. (1996) Structure and mechanism of DNA topoisomerase II. Nature, 379, 225–232. [DOI] [PubMed] [Google Scholar]
- Berger J.M., Fass,D., Wang,J.C. and Harrison,S.C. (1998) Structural similarities between topoisomerases that cleave one or both DNA strands. Proc. Natl Acad. Sci. USA, 95, 7876–7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic D., Grazulis,S., Siksnys,V. and Huber,R. (1996) Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 Å resolution. J. Mol. Biol., 255, 176–186. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cheng X., Balendiran,K., Schildkraut,I. and Anderson,J. (1994) Structure of PvuII endonuclease with cognate DNA. EMBO J., 13, 3927–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colandene J.D. and Topal,M.D. (1998) The domain organization of NaeI endonuclease: separation of binding and catalysis. Proc. Natl Acad. Sci. USA, 95, 3531–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M. and Topal,M.D. (1989) DNA and spermidine provide a switch mechanism to regulate the activity of restriction enzyme NaeI. Proc. Natl Acad. Sci. USA, 86, 9707–9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deibert M., Grazulis,S., Janulaitis,A., Siksnys,V. and Huber,R. (1999) Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 Å resolution. EMBO J., 18, 5805–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol., 276, 523–530. [PubMed] [Google Scholar]
- Doublie S., Tabor,S., Long,A.M., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- Harrison S.C. (1991) A structural taxonomy of DNA-binding domains. Nature, 353, 715–719. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Holtz J.K. and Topal,M.D. (1994) Location of putative binding and catalytic sites of NaeI by random mutagenesis. J. Biol. Chem., 269, 27286–27290. [PubMed] [Google Scholar]
- Horton N.C., Newberry,K.J. and Perona,J.J. (1998) Metal ion-mediated substrate-assisted catalysis in type II restriction endonucleases. Proc. Natl Acad. Sci. USA, 95, 13489–13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo K. and Topal,M.D. (1995) DNA topoisomerase and recombinase activities in NaeI restriction endonuclease. Science, 267, 1817–1820. [DOI] [PubMed] [Google Scholar]
- Jo K. and Topal,M.D. (1998) Step-wise DNA relaxation and decatenation by NaeI-43K. Nucleic Acids Res., 26, 2380–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck J.L. and Berger,J.M. (1999) Enzymes that push DNA around. Nature Struct. Biol., 6, 900–902. [DOI] [PubMed] [Google Scholar]
- Kim Y., Grables,J.C., Love,R., Greene,P.J. and Rosenberg,J.M. (1990) Refinement of EcoRI endonuclease crystal structure: a revised protein chain tracing. Science, 249, 1307–1309. [DOI] [PubMed] [Google Scholar]
- Kostrewa D. and Winkler,F.K. (1995) Mg2+ binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 Å resolution. Biochemistry, 34, 683–696. [DOI] [PubMed] [Google Scholar]
- Kovall R.A. and Matthews,B.W. (1997) Toroidal structure of λ-exonuclease. Science, 277, 1824–1827. [DOI] [PubMed] [Google Scholar]
- Kovall R.A. and Matthews,B.W. (1998) Structural, functional, and evolutionary relationships between λ-exonuclease and the type II restriction endonucleases. Proc. Natl Acad. Sci. USA, 95, 7893–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R.A. and Matthews,B.W. (1999) Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol., 3, 578–583. [DOI] [PubMed] [Google Scholar]
- Krüger D.H., Barcak,G.J., Reuter,M. and Smith,H.O. (1988) EcoRII can be activated to cleave refractory DNA recognition sites. Nucleic Acids Res., 16, 3997–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeda D.P. (1986) Transfer of ‘Nocardia aerocolonigenes’ () Pridham 1970 into the genus Saccharothrix Labeda, Testa, Lechevalier, and Lechevalier 1984 as Saccharothrix aerocolonigenes sp. nov. Int. J. Syst. Bacteriol., 36, 109–110. [Google Scholar]
- Lima C.D., Wang,J.C. and Mondragon,A. (1994) Three dimensional structure of the 67K N-terminal fragment of E.coli DNA topoisomerase I. Nature, 367, 138–146. [DOI] [PubMed] [Google Scholar]
- Martin A.M., Sam,M.D., Reich,N.O. and Perona,J.J. (1999) Structural and energetic origins of indirect readout in site-specific DNA cleavage by a restriction endonuclease. Nature Struct. Biol., 6, 269–279. [DOI] [PubMed] [Google Scholar]
- McClarin J.A., Frederick,C.A., Wang,B.C., Greene,P., Boyer,H.W., Grable,J. and Rosenberg,J.M. (1986) Structure of the DNA–EcoRI endonuclease recognition complex at 3 Å resolution. Science, 234, 1526–1540. [DOI] [PubMed] [Google Scholar]
- Nelson H.C.M. (1995) Structure and function of DNA-binding proteins. Curr. Opin. Genet. Dev., 5, 180–189. [DOI] [PubMed] [Google Scholar]
- Newman B.J. and Grindley,N.D.F. (1984) Mutants of the γδ resolvase: a genetic analysis of the recombination function. Cell, 38, 463–469. [DOI] [PubMed] [Google Scholar]
- Newman M., Strelecka,T., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1995) Structure of BamHI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science, 269, 656–663. [DOI] [PubMed] [Google Scholar]
- Newman M., Lunnen,K., Wilson,G., Greci,J., Schildkraut,I. and Philips,S.E.V. (1998) Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence. EMBO J., 17, 5466–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oller A.R., Vanden Broek,W., Conrad,M. and Topal,M.D. (1991) Ability of DNA and spermidine to affect the activity of restriction endonucleases from several bacterial species. Biochemistry, 30, 2543–2549. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collection in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pabo C.O. and Sauer,R.T. (1992) Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem., 61, 1053–1095. [DOI] [PubMed] [Google Scholar]
- Pingoud A. and Jeltsch,A. (1997) Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem., 246, 1–22. [DOI] [PubMed] [Google Scholar]
- Reuter M., Kupper,D., Pein,C., Petrusyte,M., Siksnys,V., Frey,B. and Krüger,D.H. (1993) Use of specific oligonucleotide duplexes to stimulate cleavage of refractory DNA sites by restriction endonucleases. Anal. Biochem., 209, 232–237. [DOI] [PubMed] [Google Scholar]
- Roberts R.J. and Macelis,D. (2000) REBASE—restriction enzymes and methylases. Nucleic Acids Res., 28, 306–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S.C., Shields,G.C. and Steitz,T.A. (1991) Crystal structure of a CAP–DNA complex: the DNA is bent by 90°. Science, 253, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Shinobu R. and Kawato,M. (1960) On Streptomyces aerocolonigenes nov. sp., forming secondary colonies on the aerial mycelia. Bot. Mag. (Tokyo), 73, 212–216. [Google Scholar]
- Taron C.H., Van Cott,E.M., Wilson,G.G., Moran,L.S., Slatko,B.E., Hornstra,L.J., Benner,J.S., Kucera,R.B. and Guthrie,E.P. (1995) Cloning and expression of the NaeI restriction endonuclease-encoding gene and sequence analysis of the NaeI restriction-modification system. Gene, 155, 19–25. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated structure solution for MIR and MAD. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal M.D. and Conrad,M. (1993) Changing endonuclease EcoRII Tyr308 to Phe abolishes cleavage but not recognition: possible homology with the Int-family of recombinases. Nucleic Acids Res., 21, 2599–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa S.E., Muto,T., Kawate,T., Jingami,H., Kunishima,N., Ariyoshi,M., Kohda,D., Nakagawa,M. and Morikawa,K. (1999) Crystallographic and functional studies of very short patch repair endonuclease. Mol. Cell, 3, 621–628. [DOI] [PubMed] [Google Scholar]
- Venclovas C., Timinskas,A. and Siksnys,V. (1994) Five-stranded β-sheet with two α-helices: a structural link between restriction endonucleases EcoRI and EcoRV. Proteins, 20, 279–282. [DOI] [PubMed] [Google Scholar]
- Viadiu H. and Aggarwal,A.K. (1998) The role of metals in catalysis by the restriction endonuclease BamHI. Nature Struct. Biol., 5, 910–916. [DOI] [PubMed] [Google Scholar]
- Wilson G.G. (1991) Organization of restriction-modification systems. Nucleic Acids Res., 19, 2539–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler F.K. et al. (1993) The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA. EMBO J., 12, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.C. and Topal,M.D. (1992) Nonidentical DNA-binding sites of endonuclease NaeI recognize different families of sequences flanking the recognition site. Biochemistry, 31, 9657–9664. [DOI] [PubMed] [Google Scholar]
- Yang C.C., Baxter,B.K. and Topal,M.D. (1994) DNA cleavage by NaeI: protein purification, rate-limiting step, and accuracy. Biochemistry, 33, 14918–14925. [DOI] [PubMed] [Google Scholar]