Abstract
In contrast to yeast RAD51, mammalian mRAD51 is an essential gene. Its role in double strand break (DSB) repair and its consequences on cell viability remain to be characterized precisely. Here, we used a hamster cell line carrying tandem repeat sequences with an I-SceI cleavage site. We characterized conservative recombination after I-SceI cleavage as gene conversion or intrachromatid crossing over associated with random reintegration of the excised reciprocal product. We identified two dominant-negative RAD51 forms that specifically inhibit conservative recombination: the yeast ScRAD51 or the yeast–mouse chimera SMRAD51. In contrast, the mouse MmRAD51 stimulates conservative recombination. None of these RAD51 forms affects non-conservative recombination or global DSB healing. Consistently, although resistance to γ-rays remains unaffected, MmRAD51 stimulates whereas ScRAD51 or SMRAD51 prevents radiation-induced recombination. This suggests that mRAD51 does not significantly affect the global DSB repair efficiency but controls the classes of recombination events. Finally, both ScRAD51 and SMRAD51 drastically inhibit spontaneous recombination but not cell proliferation, showing that RAD51-dependent spontaneous and DSB-induced conservative recombination can be impaired significantly without affecting cell viability.
Keywords: double strand break repair/homologous recombination/mammalian cells/RAD51/trans-species dominant-negative allele
Introduction
A DNA double strand break (DSB) is a lesion that can be produced by genotoxic agents such as ionizing radiation or that can occur during physiological processes such as yeast meiosis (Sun et al., 1989). Repair of DSBs is essential to ensure genome integrity and cell viability, but may also result in genome rearrangements. Two major processes can repair DSBs: non-homologous end-joining (NHEJ) or a homology-directed (HD) process that takes advantage of a homologous sequence to repair the DSB (Liang et al., 1998; for reviews see Baumann and West, 1998; Kanaar and Hoeijmakers, 1998). Actually an HD process refers to two independent pathways: the non-conservative single strand annealing (SSA) process that occurs between direct repeat sequences (Lin et al., 1984), and the conservative homologous recombination, initiated by a DNA strand invasion step. In yeast, conservative recombination is thus dependent on ScRAD51. In contrast, SSA is RAD51 independent (Ivanov et al., 1996). Thus, in mammalian cells, the comprehension of DSB repair regulation presupposes the precise determination of the different pathways involved.
The search for RAD51 orthologues has led to the identification of a growing number of mammalian RAD51 homologues (for a review see Thacker, 1999). This suggests either redundancy of functions or the splitting up of ScRAD51’s role on several mammalian genes, or an evolutionary divergence of function for some of the homologues. In this context, the mammalian mRAD51 presents both similarities to and differences from ScRAD51, and its exact role in DSB repair remains to be established. Human Rad51 is able to promote DNA strand exchange in vitro, but less efficiently and with some biochemical differences compared with the yeast protein (Baumann et al., 1996; Gupta et al., 1997; Benson et al., 1998). In vivo, chromosome breakages and a decrease in sister chromatid exchanges have also been reported prior to cell death in rad51–/– depleted DT40 cells, a chicken immortalized B-cell line harbouring a particularly high frequency of spontaneous gene conversion (Sonoda et al., 1998, 1999). Moreover, mouse Rad51 protein was shown to associate with mouse meiotic chromosomes and to relocalize to nuclear foci in cells treated with genotoxic agents (Haaf et al., 1995; Barlow et al., 1997). However, contrasting with the situation in yeast and bacteria, mRAD51 has been described to be involved in cell proliferation and is an essential gene in vertebrates since null mutants are not viable (Tsuzuki et al., 1996; Sonoda et al., 1998). Moreover, mRAD51 participates, via BRCA2, in the regulation of p53 transactivation activity (Marmorstein et al., 1998). Since the transactivation activity of p53 is involved in cell cycle control but is not correlated with the role of p53 in recombination (Dudenhoffer et al., 1999; Saintigny et al., 1999), this result may suggest the participation of mRAD51 in the cell cycle checkpoint controlled by p53 transactivation activity. Thus, chromosomal aberrations observed in rad51–/– depleted cells could also be explained by cell cycle defects.
Several hypotheses could account for these apparent paradoxes. The first hypothesis is that mRAD51 controls one DSB repair pathway essential for cell viability, as proposed for the chicken DT40 lines (Sonoda et al., 1998, 1999; Morrison et al., 1999). The second hypothesis is that mRAD51 possesses two distinct roles: a non-essential role controlling DSB repair and another essential for cell viability. A third hypothesis is that mRAD51 is involved in an essential function but not in homologous recombination, while other homologues of RAD51 would be required for homologous recombination. This latter hypothesis is based on the facts that a growing number of RAD51 homologues are being reported and that some of them, such as XRCC2 and XRCC3, are involved in HD DSB repair (Johnson et al., 1999; Pierce et al., 1999).
To determine whether mRAD51 actually acts on DSB repair in mammalian cells and to characterize its putative role precisely, we used the CHO cell line DRA10 carrying a recombinant substrate with a unique site for the rare cutting endonuclease I-SceI (Liang et al., 1998). We show that overexpression of mouse MmRAD51 does not substantially stimulate the overall DSB repair but specifically stimulates the conservative recombination DSB repair pathway. More importantly, we also identified forms of RAD51 (the yeast ScRAD51 or the yeast–mouse chimera SMRAD51) that specifically inhibit DSB-induced conservative recombination as well as spontaneous recombination. None of the RAD51 forms used here affect SSA. These results indicate that mRAD51 participates in DSB repair, does not modify the global efficiency of DSB repair but acts on the balance controlling the different classes of events: conservative versus non-conservative recombination. Finally, our results show that it is possible to decrease the efficiency of RAD51 DSB repair and RAD51 spontaneous recombination substantially without affecting cell viability and proliferation.
Results
Strategy and cell lines used
In order to define the DSB repair pathway controlled by mRAD51 precisely, we measured the impact of the overexpression of different forms of RAD51 on recombination induced either by an acute DSB targeted to the recombination substrate or by ionizing radiation. It has been shown that the overexpression of mRAD51 in mammalian cells results in an increase in spontaneous recombination between intrachromosomal repeat sequences (Vispé et al., 1998; Arnaudeau et al., 1999; Huang et al., 1999). However, it has been argued that overexpression of RAD51 interferes with DNA metabolism and results in DNA lesions that could induce recombination, as has been pointed out (Huang et al., 1999). Importantly, the role of mRAD51 in DSB repair and the putative pathways involved have not been addressed in mammalian cells.
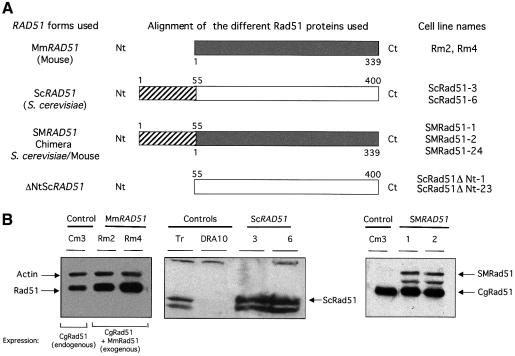
In addition, in an attempt to identify a dominant-negative RAD51 form, we expressed ScRAD51 from Saccharomyces cerevisiae. Trans-species dominant- negative effects have been described between RAD52 from Kluyveromyces lactis and S.cerevisiae (Milne and Weaver, 1993). More specifically, the active species for recombination is the RecA/Rad51 nucleoprotein filament (Radding, 1991). Alignment of mouse MmRad51 and ScRad51 shows that the yeast protein is longer than that of the mouse, with a block of 55 additional amino acids at the N-terminal extremity (Figure 1A). Furthermore, ScRAD51 interacts with the human RAD51 and XRCC3 (Schild et al., 2000). Our hypothesis was that ScRAD51 may act as a dominant-negative allele in mammalian cells by interacting with endogenous mRAD51, leading to the poisoning of the mRAD51 nucleoprotein filament via the extra N-terminal part of the yeast protein. Another possibility could be that ScRAD51 would lead to the titration of the endogenous recombination complex. To check these hypotheses, we also expressed the chimera SMRad51 composed of the 55 N-terminal amino acids from ScRad51 fused to the entire MmRad51, and ΔNtScRad51 corresponding to ScRad51 with its 55 N-terminal amino acids deleted (see Figure 1A).
Fig. 1. Overexpression of the different RAD51 genes in hamster cells. (A) Structure of the RAD51 genes used. The boxes correspond to the homologous region between the different RAD51 genes: grey, mouse MmRAD51; white, yeast ScRAD51. ScRAD51 is longer at the N-terminal part. The white box corresponds to the region homologous to MmRAD51 and the hatched box to the extra sequence. The numbers correspond to the positions of the amino acids. The numbers in black correspond to the yeast amino acids and the numbers in grey correspond to the mouse amino acids. The chimeric protein SMRad51 was constructed with the N-terminal part of ScRad51 (hatched box) fused to the entire MmRad51 (white box). ΔNtScRad51 corresponds to ScRad51 with 55 N-terminal amino acids deleted. These proteins are expressed in the CHO-DRA10 line, and the corresponding derivative lines are listed on the right. (B). Overexpression of exogenous RAD51. Protein extracts were obtained from stable transfectants. The expression of MmRAD51 and SMRAD51 was verified by western blotting using an antibody raised against the human Rad51 protein and normalized with an anti-actin antibody. Rm2 and 4 correspond to two independent stable transfectants overexpressing MmRAD51. SMRad51-1 and SMRad51-2 correspond to two independent stable transfectants overexpressing the fusion SMRAD51. Cm3 is a control clone corresponding to the CHO-DRA10 line transfected with the empty pCDNA3.1puro plasmid. Expression of ScRAD51 is visualized by western blotting using an anti-ScRad51 antibody. The first lane corresponds to transient expression (Tr) of ScRAD51 (used as control). The second lane corresponds to extracts from mock-transfected cells (CHO-DRA10). The third and fourth lanes correspond to two independent clones with stable expression of ScRAD51. MmRad51 is Rad51 from Mus musculus; ScRad51 is Rad51 from S.cerevisiae; CgRad51 is Rad51 from Cricetellus griseus; and SMrad51 is a chimeric fusion protein MmRad51–ScRad51.
These different RAD51 forms are expressed in the parental CHO-DRA10 line (Liang et al., 1998), and the names of the derivative cell lines are listed in Figure 1A. Figure 1B shows the expression of the endogenous and exogenous RAD51s, verified by western blotting. Expression of ΔNtScRAD51 has been verified by RT–PCR (data not shown).
Characterization of the DSB-induced recombination events
The CHO-DRA10 cell line and its derivatives carry a unique recombination substrate with the cleavage site for the rare-cutting endonuclease I-SceI (see Figure 2). Transient expression of I-SceI efficiently produces a DSB in the recombination substrate (Liang et al., 1998).
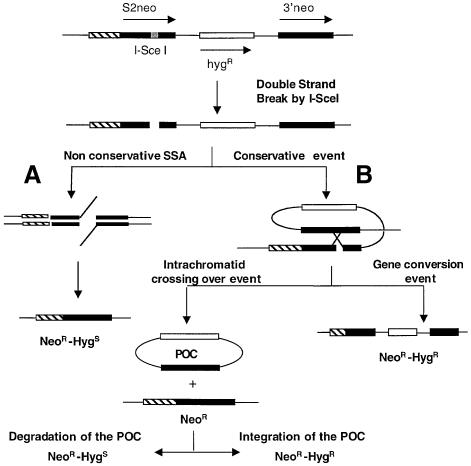
Fig. 2. Strategy used to measure DSB-induced recombination events. After the formation of a DSB induced by I-SceI, two processes can compete. (A) The non-conservative SSA that produces NeoR and hygromycin-sensitive recombinants. (B) Homologous recombination events leading to gene conversion either with or without crossing over. The products of gene conversion events are resistant to both G418 and hygromycin (NeoR–HygR). Intrachromatid crossing over leads to the formation of POCs. If the POCs are eliminated, the products are only NeoR; if the POC is reintegrated into the genome, it could result in a NeoR–HygR colony. Thus, double resistance (NeoR–HygR) scores only conservative recombination events. Not shown are unequal sister chromatid exchange events in which the HygR segregates from the NeoR gene.
Single-drug resistance to G418 (NeoR) monitors recombinant clones arising by an HD recombination process, both conservative and non-conservative (SSA) events (Figure 2). Double resistance to both G418 (NeoR) and hygromycin (HygR) monitors only conservative events due to gene conversion not associated with crossing over or associated with crossing over, but with a faithful reintegration of the excised HygR sequence (Figure 2B). NeoR–Hygs monitors SSA events plus crossing over with loss of the pop out circle (POC) (Figure 2A).
To demonstrate the occurrence of the recombination products predicted in Figure 2, we analysed the molecular structure of the recombination locus from NeoR–HygR clones by Southern blotting (Figure 3A). We analysed 11 NeoR–HygR recombinant clones, seven from the control line Cm3 and four from the MmRAD51-overexpressing line Rm2 (Figure 3). With the S2neo sequence probe (probe A), nine clones show the gene conversion 4 kb band and two recombinant clones show the 1.1 kb deletion event band (Figure 3B), despite the fact that these two clones exhibit a hygromycin-resistant phenotype. We thus probed the same filters with the hygromycin resistance gene sequence (probe B). The structure of the nine gene conversion events previously identified is confirmed, since they show the 4 kb restriction band with this new probe (Figure 3C). In the two deleted clones, however, the presence of the hygromycin resistance gene sequence is revealed but at a different location (Figure 3C), suggesting a random integration after a crossing over event. These results show that NeoR–HygR clones actually score gene conversion and some crossing over events, both initiated by strand invasion (Szostak et al., 1983). In addition, these results demonstrate the existence of the predicted DNA fragment excised by an intrachromatid crossing over (see Figure 2B), followed by its random integration into the genome.
Fig. 3. (A) Restriction pattern of recombinant clones induced by an I-SceI-generated DSB. Probe A corresponds to the S2neo promoter sequence and probe B to part of the hygromycin-resistant gene, located between the two neo cassettes. The corresponding expected XhoI–HindIII sizes (with both probes) and the corresponding resistance phenotype are indicated. POC: pop out circle (see Figure 2). (B) Southern blot analysis with probe A. The sizes of the bands are indicated on the sides of the panels. Left panel: seven independent NeoR–HygR clones (numbered on the top of the panel) from the control cell line Cm3. Right panel: four independent NeoR–HygR clones (numbered on the top of the panel) from the Rm2 cell line, overexpressing MmRAD51. (C) The same filters as in (B) but hybridized to probe B, characteristic of the intervening sequence.
RAD51 specifically affects conservative recombination events in mammalian cells
We measured here the impact of the overexpression of the different RAD51 forms (Figure 1) on recombination induced by an I-SceI DSB, targeted to the recombination substrate.
We first verified that the transfection efficiency of the I-SceI expression vector was equivalent in the control lines and the overexpressing RAD51 cell lines (data not shown). We also measured the same 100- to 1000-fold increase in the frequency of G418-resistant colonies (total recombinant clones) following transient transfection of the I-SceI expression vector (Liang et al., 1998). As compared with the control cell line (Cm3), increases in the frequency of G418-resistant clones by factors of 1.8–2.6 are found in lines expressing MmRAD51 and decreases in the frequency of G418-resistant clones by factors of 1.7–2.4 in lines expressing ScRAD51 or SMRAD51 (Table I).
Table I. Effect of different Rad51 proteins on DSB-induced recombination.
| Cell line | Mean NeoR frequency (× 10–3)a | Stimulation factorb | Mean NeoR–HygR frequency (× 10–3)a | Stimulation factorb |
|---|---|---|---|---|
| Cm3 | 1.19 ± 0.2 | 0.38 ± 0.1 | ||
| Rm2 | 3.09 ± 0.7 | 2.6 | 2 ± 0.4 | 5.3 |
| Rm4 | 2.1 ± 0.9 | 1.8 | 1.59 ± 0.9 | 4.2 |
| Inhibition factorb | Inhibition factorb | |||
| ScRad51-3 | 0.64 ± 0.4 | 1.8 | 0.008 ± 0.005 | 47.5 |
| SMRad51-1 | 0.71 ± 0.1 | 1.7 | 0.009 ± 0.007 | 42.2 |
| SMRad51-2 | 0.5 ± 0.2 | 2.4 | 0.003 ± 0.002 | 126.7 |
aMean values from three independent experiments.
bCompared with the Cm3 cell line control.
The impact of overexpression of the different RAD51 forms is much stronger on the frequency of NeoR–HygR recombinants. Indeed, overexpression of MmRAD51 stimulates the frequency of NeoR–HygR colonies by 4–5 times compared with the control line Cm3 (Table I). In contrast, the frequency of NeoR–HygR colonies is decreased 48 times by expression of ScRAD51. The chimera SMRAD51 also exhibits a pronounced dominant-negative effect since inhibition of the occurrence of NeoR–HygR colonies is decreased 42- to 127-fold compared with the control line Cm3 (Table I).
The percentage of conservative events (double-resistant NeoR–HygR colonies) relative to all the recombinant colonies (NeoR alone) was examined (Figure 4). Since this value is normalized to the frequency of NeoR colonies (representing the whole recombinant population), the calculation is based on an internal standard and is independent of transfection and cleavage efficiencies. In the Cm3 control line, the frequency of conservative events (NeoR–HygR colonies) is 27%, and thus the non-conservative events represent 73% of the total recombinant colonies. Comparable ratios are obtained with the parental CHO-DRA10 line (Liang et al., 1998). In the lines overexpressing the MmRAD51 gene, the relative proportion of NeoR–HygR colonies rises to 65–72% (Figure 4). In contrast, the percentage of NeoR–HygR colonies falls to 2% in the line expressing ScRAD51, and to 1.6 or 0.9% in lines expressing SMRAD51 (Figure 4). The expression of the deleted ΔNtScRAD51 has no effect on the distribution of the different events, compared with the control lines (Figure 4). These results demonstrate a dominant-negative effect of ScRAD51 and of the chimera SMRAD51 with respect to the homologous recombination initiated by strand invasion. In addition, the dominant-negative effect requires the 55 N-terminal amino acids of the ScRAD51 product.
Fig. 4. Effect of the overexpression of MmRAD51, ScRAD51 or SMRAD51 on the frequency of conservative recombination events. Control refers to the parental line transfected with empty expression vector (Cm3). MmRAD51 refers to two independent transfectants overexpressing MmRAD51; ScRAD51 refers to one transfectant expressing yeast ScRAD51; SMRad51-1 and SMRad51-2 refer to two independent clones expressing the chimera SMRAD51; ScRad51ΔNt-1 and ScRad51ΔNt-23 refer to two independent clones expressing the deleted ΔNtScRAD51. Values correspond to the percentage of double-resistant (NeoR–HygR) colonies (black boxes) and single-resistant (NeoR) colonies (grey boxes). The percentage of conservative recombination events is calculated from the ratio of the number of NeoR–HygR clones to the NeoR clone frequency. The percentages are indicated on the top of the corresponding histograms.
The present data agree completely with the view that DSBs can be repaired by NHEJ, by a conservative strand invasion mechanism or by SSA. Indeed, expression of MmRAD51, ScRAD51 or SMRAD51 only very moderately affects the total frequency of recombinants, indicating that the ratio of NHEJ to HD repair is not modified significantly. In addition, overexpression of MmRAD51 specifically increases the occurrence of conservative events, whereas overexpression of ScRAD51 or of the chimera SMRAD51 prevents conservative events. Thus, the different RAD51 forms do not affect the SSA process itself, but modify the ratio of conservative events to SSA.
RAD51 controls radiation-induced recombination but not radiation resistance
In the above experiments, RAD51 does not substantially affect the total efficiency of DSB healing but rather the ratio of the different recombination classes. Since DSB is one of the main lethal lesions induced by ionizing radiation, we measured the impact of expression of different RAD51 forms on radiation resistance and on radiation-induced recombination.
None of the different RAD51 forms modify the radiation resistance of the recipient cell lines in the dose range tested (Figure 5A). These results are consistent with our previous I-SceI experiments showing that mRAD51 does not affect the efficiency of global DSB healing but controls the classes of recombination events. Indeed, radiation-induced recombination is strongly dependent on the type of exogenous RAD51 expressed: MmRAD51 expression stimulates recombination frequency 10- to 20-fold after a dose of 6 Gy, whilst SMRAD51 as well as ScRAD51 completely abolish the induction of radiation-induced recombination (Figure 5B). These results show that in an asynchronous cell population of mammalian cells, radiation resistance is a RAD51-independent process whereas radiation-induced recombination is a RAD51-dependent process.
Fig. 5. Effect of RAD51 on radiation resistance (A) and on radiation-induced recombination (B). Cells were irradiated at the doses indicated. Controls correspond to the parental CHO-DRA10 line and to Cm3, which is CHO-DRA10 transfected with the empty expression vector. Rm2 and Rm4 are two independent clones overexpressing MmRAD51. ScRad51-3 and ScRad51-6 are two independent clones expressing ScRAD51. Radiation-induced recombination (B): the values correspond to the number of NeoR in 106 surviving irradiated cells, following subtraction of the number of NeoR in 106 non-irradiated cells.
Spontaneous recombination is affected by ScRAD51 and SMRAD51
RAD51 is an essential gene in non-irradiated mammalian cells. It has been proposed that spontaneous recombination controlled by mRAD51 would be essential for cell viability by repairing spontaneous damage occurring during replication; this would also be different from yeast. However, the above results show that RAD51 recombinational DSB repair can be decreased substantially without modifying the cell viability after radiation. Taking into account our results, the proposed hypothesis should thus consider that spontaneous and DSB-induced recombination are two separable mechanisms involving mRAD51. We thus tested whether spontaneous recombination can also be inhibited by ScRAD51 and SMRAD51, as is the case for DSB-induced recombination.
Overexpression of mRAD51 has previously been reported to increase spontaneous recombination (Vispé et al., 1998; Arnaudeau et al., 1999; Huang et al., 1999). However, none of these studies used a fluctuation analysis to measure the process. In order to measure spontaneous recombination in an exponentially growing population, we performed fluctuation analysis using the Luria and Delbruck assay (Luria and Delbruck, 1943), adapted by Capizzi and Jameson (1973). As expected, we confirmed that MmRAD51 stimulates spontaneous recombination 3- to 4-fold per cell per generation (Figure 6A).
Fig. 6. Effect of the overexpression of the different RAD51 forms on spontaneous recombination. (A) MmRAD51 stimulates the spontaneous recombination rate measured by fluctuation analysis using the Luria and Delbruck test (Luria and Delbruck, 1943). The names of the different lines are indicated under the histograms. DRA10, parental line; Cm3 (control line), DRA10 transfected with empty expression vector; Rm2 and Rm4 correspond to two independent clones overexpressing MmRAD51. The number of independent cultures for each line is reported under the histograms. (B) ScRAD51 and SMRAD51 inhibit the frequency of spontaneous recombination. A recombination rate cannot be calculated because most of the cultures do not contain any recombinant. The frequency of recombination reported here corresponds to the sum of the recombinants from all the independent cultures in relation to the total number of cells summed from all the independent cultures. DRA10 is the parental line; ScRad51-3 and ScRad51-6 correspond to two independent clones expressing ScRAD51; SMRad51-1, SMRad51-2 and SMRad51-24 refer to three independent clones expressing the chimera SMRAD51.
In contrast to MmRAD51, ScRAD51 and the chimera SMRAD51 strongly decrease the spontaneous recombination frequency in mammalian cell lines, in an interspecies dominant-negative manner (Figure 6B). The recombination inhibition is so pronounced that the recombination rate cannot be calculated by the Luria and Delbruck test. Indeed, most of the cultures contain no recombinant colonies. Consequently, ScRAD51 as well as SMRAD51 also inhibit spontaneous recombination in growing cells, in a dominant-negative manner.
Finally, despite the fact that spontaneous recombination is almost totally defective, neither the generation time, the plating efficiency nor the cell cycle measured by flow cytometry (data not shown) are modified in the different lines expressing ScRAD51 or SMRAD51, showing that cell viability and proliferation are unaffected in these lines.
Discussion
The different lines devised here derive from the same parental line. Thus, recombination frequencies are calculated for one copy of substrate located at the same locus in each cell line. Using these lines, we have determined the precise pathway involving mRAD51 for DSB repair in mammalian cells: it acts specifically on conservative recombination and does not affect non-conservative SSA. This phenotype is similar to that of ScRAD51 in yeast, despite the numerous differences between the yeast and mammalian RAD51 protein products. However, an important difference from the yeast model is that in mammalian cells, mRAD51 DSB repair can be decreased substantially without significant effects on the global DSB repair efficiency. Indeed, in yeast, alteration of the RAD51 pathway leads to a drastic decrease in the global DSB repair efficiency. In mammalian cells, a balanced regulation maintains the total level of DSB repair efficiency, but modifies the class of recombination events.
Southern blot analysis of DSB-induced NeoR–HygR clones shows the existence of both gene conversion and crossing over events, the former being detected more frequently in the system used here. The restriction patterns of some clones are compatible with an intrachromatid crossing over followed by a random reintegration of the excised DNA. Although the putative circular nature of the intermediate excised DNA remains to be established, the present data demonstrate the genome rearrangement predicted by the intrachromatid crossing over model (see Figure 2). Such processes have important biological consequences with regard to genome stability/variability. For example, POCs created by intrachromatid crossing over in rDNA repeats are involved in cell ageing in yeast (Sinclair and Guarente, 1997). In addition, random integration of an excised fragment generated by a DSB-induced intrachromatid crossing over can inactivate a recipient integration locus. Taking into account the high number of homologous repeat sequences dispersed throughout the genome, the process described here could correspond to an actual mutagenic mechanism, particularly after treatments generating DSBs, such as ionizing radiation.
When inducing one DSB, targeted to one repeat of the duplication, the overexpression of MmRAD51 only slightly stimulates the frequency of total recombinant clones (NeoR). However, the ratio of NeoR–HygR to NeoR clones is increased several-fold. These results demonstrate that MmRAD51 specifically increases conservative recombination events and not SSA events (most of the NeoR clones). In yeast, ScRAD51 acts in a complex comprising the components of the RAD52 epistasis group (Milne and Weaver, 1993; Hays et al., 1995; Johnson and Symington, 1995; Rattray and Symington, 1995). In mammalian cells, homologues of most of these genes have been described. In addition, mRAD51 interacts with another set of proteins without homologues in yeast, such as the tumour suppressor and cell cycle control proteins p53, BRCA1 and BRCA2 (Sturzbecher et al., 1996; Buchhop et al., 1997; Mizuta et al., 1997; Scully et al., 1997; Marmorstein et al., 1998). In line with this, p53 is involved in homologous recombination independently of its role in G1/S transition and of its transactivation activity (Dudenhoffer et al., 1999; Saintigny et al., 1999), a result compatible with a role for p53 in the homologous recombination process. In addition, BRCA1 has been shown to be involved in DSB repair in mammalian cells (Moynahan et al., 1999). Taken together, these data suggest that mRAD51 acts in one (or several) highly elaborated complex(es). This raises the question as to how overexpression of only one component of such (a) complex(es) can stimulate the whole recombination process. The present results suggest that RAD51 is a limiting factor for homologous recombination in mammalian cells. Recombination stimulation by overexpression of RAD52, another component of the complex, has also been described in mammalian cells (Park, 1995). However, it is not known whether both conservative recombination and SSA events are stimulated since RAD52 also promotes SSA (Fishman-Lobell et al., 1992; Mortensen et al., 1996; Van Dyck et al., 1999). Thus, it cannot be excluded that this observation in mammalian cells refers to stimulation of SSA events. Consistent with this hypothesis, it has been suggested that Rad52 and Ku proteins compete for binding to DSBs, leading to the channelling of the DSB repair to an HD DSB repair or to an NHEJ process (Van Dyck et al., 1999). Our results suggest that RAD51 does not substantially affect the channelling of the NHEJ versus the HD repair process, but that inside the HD pathway (the RAD52 pathway), RAD51 may favour the strand invasion over the SSA recombination pathway. This observation is consistent with the fact that mRAD51 does not modify radiation sensitivity since the global efficiency of DSB healing remains essentially unaffected. Moreover, the fact that the status of RAD51 affects the extent of radiation-induced recombination is compatible with the hypothesis of a channelling role for mRAD51 towards the conservative recombination pathway. The simplest explanation would be that RAD51 constitutes a limiting factor for the homologous recombination pathway. RAD52 would bind the DSB and would predominantly channel repair to SSA, the most efficient HD process in these experiments (Liang et al., 1998; this study). Alternatively, RAD52 could load mRAD51 on the broken DNA, resulting in the channelling of DSB repair towards a conservative homologous recombination process (Figure 7). With such a hypothesis, increasing the intracellular amount of mRAD51 should favour the channelling of the repair to strand invasion conservative events, as is actually the case in our experiments.
Fig. 7. Role of RAD51 in DSB repair in mammalian cells. On a DSB, Ku and RAD52 compete to process the DSB. Ku orientates DSB repair towards NHEJ. Conversely, RAD52 orientates repair to an HD process that can be either SSA or homologous recombination. In the absence of RAD51, SSA takes place. This is the most frequent reaction in the parental lines. If mRAD51 is present, it can bind to RAD52 and channel the reaction to homologous recombination initiated by strand invasion.
Expression of ScRAD51 specifically inhibits conservative events in mammalian cells. The 55 N-terminal amino acids of the protein appear to be essential for this specific dominant-negative effect. Indeed, ScRad51 lacking these N-terminal amino acids (ΔNtScRad51) loses its dominant-negative effect. In contrast, the fusion of these 55 amino acids to mouse MmRad51 (SMRad51) confers a specific dominant-negative effect to the chimera. Our hypothesis was that ScRAD51 molecules would be incorporated into the mRAD51 single-stranded DNA nucleoprotein filament, generally considered as the active intermediate in the homologous recombination process (Radding, 1989); indeed, ScRAD51 has been shown to interact with human RAD51 (Schild et al., 2000). However, our hypothesis was also that the divergences between the two kinds of RAD51 molecules would then poison the nucleoprotein filament and inhibit the RAD51-dependent recombination. Consistent with this hypothesis is the fact that the chimera SMRAD51 specifically inhibits recombination initiated by a strand invasion, whereas the wild-type MmRAD51 stimulates it. An alternative explanation would be that the remaining homologies between the protein products of mRAD51 and ScRAD51 are sufficient to promote interactions with some of mRAD51’s partners, but that the divergence would impair the formation of a functional recombination complex. ScRAD51 could thus titrate the components of the recombination complex. Our results do not argue in favour of this second hypothesis. Indeed, the N-terminal part of ScRad51 is essential to poison mammalian homologous recombination, but this part of the protein is absent in its mammalian orthologue; in contrast, the rest of the yeast protein (amino acids 55–400) is unable to titrate the mammalian complex, despite its strong sequence homologies to mRad51 that should favour interactions with endogenous mRad51’s partners. In addition, interactions at least with Rad52 have been reported to be species specific (Shen et al., 1996). Nevertheless, from the genetic point of view, ScRAD51 as well as SMRAD51 are dominant-negative alleles specific for homologous recombination in mammalian cells; they constitute useful and universal tools to devise cell lines specifically defective in conservative recombination without affecting cell proliferation and viability.
XRCC2 and XRCC3 proteins show homologies to Rad51 and they interact with each other and with Rad51 (Schild et al., 2000). Cell lines mutant for XRCC2 or XRCC3 are deficient in recombinational DSB repair (Johnson et al., 1999; Pierce et al., 1999). Moreover, these XRCC2 or XRCC3 mutant lines are sensitive to ionizing radiation, correlating the radiation resistance to the efficiency of recombination (Jones et al., 1987; Liu et al., 1998). In contrast, this correlation does not exist with our lines expressing the SMRAD51 (or ScRAD51) dominant-negative form. Several hypotheses could account for these apparent differences. First, our lines contain an endogenous wild-type RAD51, and one can suggest that low or undetectable levels of homologous recombination would be sufficient to maintain radiation resistance; however, radiation-induced recombination is strongly decreased. Secondly, one of the other paralogues can substitute for RAD51 for radiation resistance but not for radiation-induced recombination; they would be unable to substitute for XRCC2 or XRCC3. These two hypotheses uncouple radiation resistance and homologous recombination efficiency. A third hypothesis reconciles all the different results. This hypothesis proposes that RAD51 initiates the recombination process and that its partners, such as XRCC2 or XRCC3, facilitate the completion of the process by maturing and resolving the recombination intermediates generated by RAD51. Non-processed intermediates would result in cell toxicity. We show here that radiation-induced recombination is a RAD51-dependent pathway; thus, radiation stimulates the initiation of recombination by RAD51. The absence of XRCC2 or XRCC3 would lead to the accumulation of non-processed intermediates and would result in radiation sensitivity of the XRCC2 or XRCC3 mutant cell lines. If the initiation of homologous recombination is impaired by the dominant-negative SMRAD51, on the one hand toxic recombination intermediates would not accumulate and on the other hand DSBs would be repaired by another pathway such as NHEJ. In this hypothesis, cells should not be radiation sensitive. We do no have direct evidence that SMRAD51 acts at initiation or at a later step of strand invasion. However, our results are compatible with the channelling towards SSA or NHEJ for DSB repair. In addition, our results and the third hypothesis are fully consistent with the fact that no RAD51 mutant alleles have been isolated in the screen for radiation-sensitive mutants in CHO cell lines.
RAD51 is an essential gene in non-irradiated mammalian cells. It has been proposed that spontaneous recombination controlled by mRAD51 would be essential for cell viability by repairing spontaneous damage occurring during replication. Remarkably, the present results show that efficient mRAD51 recombination repair of DSBs induced by I-SceI or ionizing radiation is not required for cell viability. This could suggest that DSB-induced recombination and cell viability are two separable functions of mRAD51. It can be argued that the essential role of mRAD51 is spontaneous recombination. This does not agree with our results showing that both DSB-induced recombination and spontaneous recombination are stimulated by MmRAD51 and both are inhibited by ScRAD51 or SMRAD51. However, since the endogenous wild-type RAD51 is still present in our cell lines, we cannot exclude the possibility that low or undetectable levels of recombination would be sufficient to ensure its putative role in cell proliferation and viability. Nevertheless, the efficiency of the mRAD51 recombination pathway, involved in spontaneous as well as DSB-induced conservative recombination, can be decreased substantially without affecting cell proliferation and viability. However, since deletion of the RAD51 gene is lethal in mammalian cells, this could suggest that mRAD51 has other role(s), essential to cell viability. Its interactions with p53, BRCA1 and BRCA2 (Sturzbecher et al., 1996; Buchhop et al., 1997; Mizuta et al., 1997; Scully et al., 1997; Marmorstein et al., 1998) and its participation in the regulation of p53 transactivation activity (Marmorstein et al., 1998) suggest a potential role in the cell cycle checkpoint (beside potential roles in DNA repair), which could be a good candidate for this new putative essential function of RAD51. Indeed, no good structural homologues of the mRAD51 interactors p53, BRCA1 and BRCA2 have been identified in yeast; in addition, both null BRCA–/– mice and null RAD51–/– mice show embryonic lethality that can be partially rescued by inactivation of the p53 gene (Lim and Hasty, 1996; Hakem et al., 1997). Another possible essential role could be the protective effect of RAD51 against apoptosis (Huang et al., 1999). A connection between these two roles of RAD51 cannot be excluded.
A growing amount of evidence connects mRAD51 to tumorigenesis: mRAD51 interacts with tumour suppressor gene products and, in addition, mutations or loss of heterozygosity have been reported in some cancers for chromosomal loci containing BRCA1, BRCA2 and RAD51B or mRAD51’s partners RAD52, RAD54 and RAD54B (Gonzalez et al., 1999; Hiramoto et al., 1999; Matsuda et al., 1999; Schoenmakers et al., 1999). Our results show that cell viability and proliferation remain unaffected in the absence of an efficient mRAD51-dependent spontaneous and DSB-induced recombination process, but that mRAD51 participates in DSB repair by controlling the types of recombination events, i.e. conservative recombination. This function should confer an important role on mRAD51 in the maintenance of genetic stability of vegetative cells beside an essential function in cell proliferation.
Materials and methods
DNA manipulations
All DNA manipulations were performed as described (Sambrook et al., 1989).
Cells and plasmids
CHO-DRA10 cells (Liang et al., 1998) and their derivative lines were cultured at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum. NeoR clones were selected in 500 µg/ml G418, and hygromycin-resistant clones were selected in 500 µg/ml hygromycin. Transfections were performed using Fugene 6 (Boehringer, Mannheim). The SMRAD51 cDNA was constructed as follows: the N-terminal part of ScRAD51 was amplified by PCR using a 5′ primer containing an EcoRI restriction site and the beginning of the yeast gene (ccggaattcATGtctcaagaacaacat), and a 3′ primer containing the sequence corresponding to the last amino acids of the N-terminal part of ScRAD51 and the beginning of MmRAD51 cDNA up to the AvaII site (and without the first ATG). The sequence of this 3′ oligonucleotide is: gctgtggaccaaaactttcttcttccactgaagtatctgcacttgcttcaagctgcatttgcatagcgccgttggt ggcctcaatatc. The rest of the MmRAD51 sequence was amplified using a 5′ primer at the AvaII site (ttttggtccacagcctatttcaccgc) and a 3′ primer at the end of the MmRAD51 cDNA and containing a XhoI site (cggccgctcgagggagtccagtctttggcatcgccc). Double digests of the PCR products with EcoRI–AvaII and AvaII–XhoI were co-cloned in pBluescript vector digested by EcoRI–XhoI. MmRAD51, ScRAD51 and the fusion SMRAD51 cDNAs were cloned in pcDNA3.1puro plasmid. This plasmid was constructed by replacing the 1.7 kb PvuII fragment of pcDNA3.1Zeo (In Vitrogen) by the 1.4 kb PvuII–BamHI fragment from pPuro plasmid (Clontech). We used the I-SceI (HA-tagged) expression vector described by Liang et al. (1998). The deleted ΔNtScRAD51 was constructed as follows: the cDNA coding for amino acids 55–400 was amplified by PCR using an intact ScRAD51 cDNA as matrix, a 5′ primer (cggccgctcgagccaccatgtccggcgatggtggcgga) and a 3′ primer (atcaccaaatacctactcgtcttc). The amplimer was digested with XhoI and cloned in pBluescript vector. After verification of the sequence, an XhoI–EcoRV fragment was cloned in PcDNA3.1puro.
Western blot analysis
All extract preparation steps were performed at 4°C. After washing with phosphate-buffered saline (PBS), cells were suspended in lysis buffer A [25 mM Tris pH 7.5, 5 mM EDTA, 600 mM NaCl, 1 mM dithiothreitol (DTT), 0.1% NP-40, 5 µg/ml leupeptin, 2 µM pepstatin, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10% glycerol] and incubated for 40 min on ice. Extracts were centrifuged for 30 min at 15 000 g, the supernatant was retrieved and the protein concentration was determined using a Bio-Rad Protein Assay (Bio-Rad). Forty micrograms per well of the boiled samples were loaded on a 10% polyacrylamide gel in the presence of SDS. After migration, the proteins were electrotransferred onto a nitrocellulose membrane and probed with specific antibodies: anti-human Rad51 and anti-actin (Sigma). Standard procedures were used for the electrophoresis, transfer and western blotting. Antibodies were visualized using the ECL detection kit (Amersham).
Recombination measurements
Recombination after induction of a DSB. A total of 3 × 105 cells (for the Rm2, Rm4, DRA10 and Cm3 lines) and up to 1.8 × 106 cells (for the ScR and SM lines) were plated and transfected with 2 and 12 µg, respectively, of an expression vector for the I-SceI endonuclease (pCMV I-SceI). At 24 h post-transfection, G418 or G418/hygromycin selection was added. The NeoR and NeoR–HygR colony frequencies are expressed in relation to the total number of cells plated. The relative percentage of conservative recombination events is calculated as the ratio of the frequency of double-resistant NeoR–HygR clones to the frequency of Neo clones.
Recombination frequency after γ-radiation. Cells were irradiated in PBS, using a 60Co irradiator (2.5 Gy/min) at the dose indicated. After irradiation, the cells were incubated in DMEM at 37°C for 24 h. The cells were then trypsinized, counted and divided into two fractions. The first fraction was used to calculate the viability by measuring the plating efficiency. The second fraction was plated under G418 selection to measure the recombination frequency.
Fluctuation analysis for spontaneous recombination was performed as previously described (Liskay et al., 1984). For each line analysed, several independent cultures were plated and cultured to confluence. Cells were then trypsinized, counted and one fraction was used for plating efficiency estimation. The remaining cells were plated under G418 selection. The resulting number of TK+ or Neo+ clones allowed us to calculate the recombination frequency. The rate of recombination per cell per generation was calculated by using the fluctuation tests of Luria and Delbruck (Luria and Delbruck, 1943; Capizzi and Jameson, 1973).
Acknowledgments
Acknowledgements
We thank Drs D.Bishop, P.Sung, C.Radding and S.West for the anti-Rad51 antibodies, Dr M.Jasin for the CHO-DRA10 line and the I-SceI expression vector, and Drs F.Fabre and T.Morita for the yeast and mouse RAD51 cDNAs, B.Chaput for helpful and efficient technical assistance, and Drs F. Fabre, P.Radicella, C.White and members of the laboratory for helpful and stimulating discussions. S.L. is supported by an INSTN fellowship. This work was supported by Electricité de France, ARC (9822 and 9238) and ANRS.
References
- Arnaudeau C., Helleday,T. and Jenssen,D. (1999) The RAD51 protein supports homologous recombination by an exchange mechanism in mammalian cells. J. Mol. Biol., 289, 1231–1238. [DOI] [PubMed] [Google Scholar]
- Barlow A.L., Benson,F.E., West,S.C. and Hulten,M.A. (1997) Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J., 16, 5207–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. and West,S.C. (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- Baumann P., Benson,F.E. and West,S.C. (1996) Human RAD51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell, 87, 757–766. [DOI] [PubMed] [Google Scholar]
- Benson F.E., Baumann,P. and West,S.C. (1998) Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature, 391, 401–404. [DOI] [PubMed] [Google Scholar]
- Buchhop S., Gibson,M.K., Wang,X.W., Wagner,P., Sturzbecher,H.W. and Harris,C.C. (1997) Interaction of p53 with the human Rad51 protein. Nucleic Acids Res., 25, 3868–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capizzi R.L. and Jameson,J.W. (1973) A table for the estimation of the spontaneous mutation rate of cells in culture. Mutat. Res., 17, 147–148. [DOI] [PubMed] [Google Scholar]
- Dudenhoffer C., Kurth,M., Janus,F., Deppert,W. and Wiesmuller,L. (1999) Dissociation of the recombination control and the sequence-specific transactivation function of p53. Oncogene, 18, 5773–5784. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J., Rudin,N. and Haber,J.E. (1992) Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol., 12, 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Silva,J.M., Dominguez,G., Garcia,J.M., Martinez,G., Vargas,J., Provencio,M., Espana,P. and Bonilla,F. (1999) Detection of loss of heterozygosity at RAD51, RAD52, RAD54 and BRCA1 and BRCA2 loci in breast cancer: pathological correlations. Br. J. Cancer, 81, 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.C., Bazemore,L.R., Golub,E.I. and Radding,C.M. (1997) Activities of human recombination protein Rad51. Proc. Natl Acad. Sci. USA, 94, 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T., Golub,E.I., Reddy,G., Radding,C.M. and Ward,D.C. (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA, 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R., de la Pompa,J.L., Elia,A., Potter,J. and Mak,T.W. (1997) Partial rescue of Brca1 (5–6) early embryonic lethality by p53 or p21 null mutation. Nature Genet., 16, 298–302. [DOI] [PubMed] [Google Scholar]
- Hays S.L., Firmenich,A.A. and Berg,P. (1995) Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55 and Rad57 proteins. Proc. Natl Acad. Sci. USA, 92, 6925–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto T. et al. (1999) Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene, 18, 3422–3426. [DOI] [PubMed] [Google Scholar]
- Huang Y. et al. (1999) Role for caspase-mediated cleavage of Rad51 in induction of apoptosis by DNA damage. Mol. Cell. Biol., 19, 2986–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov E.L., Sugawara,N., Fishman-Lobell,J. and Haber,J.E. (1996) Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics, 142, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.D. and Symington,L.S. (1995) Functional differences and interactions among the putative RecA homologs Rad51, Rad55 and Rad57. Mol. Cell. Biol., 15, 4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.D., Liu,N. and Jasin,M. (1999) Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature, 401, 397–399. [DOI] [PubMed] [Google Scholar]
- Jones N.J., Cox,R. and Thacker,J. (1987) Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res., 183, 279–286. [DOI] [PubMed] [Google Scholar]
- Kanaar R. and Hoeijmakers,J.H. (1998) Genetic recombination. From competition to collaboration. Nature, 391, 335, 337–338. [DOI] [PubMed] [Google Scholar]
- Liang F., Han,M., Romanienko,P.J. and Jasin,M. (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.L., Sperle,K. and Sternberg,N. (1984) Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol., 4, 1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R.M., Stachelek,J.L. and Letsou,A. (1984) Homologous recombination between repeated chromosomal sequences in mouse cells. Cold Spring Harbor Symp. Quant. Biol., 49, 183–189. [DOI] [PubMed] [Google Scholar]
- Liu N. et al. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell, 1, 783–793. [DOI] [PubMed] [Google Scholar]
- Luria S.E. and Delbruck,M. (1943) Mutations of bacteria from virus sensitivity to viris resistance. Genetics, 28, 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein L.Y., Ouchi,T. and Aaronson,S.A. (1998) The BRCA2 gene product functionally interacts with p53 and RAD51. Proc. Natl Acad. Sci. USA, 95, 13869–13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M. et al. (1999) Mutations in the RAD54 recombination gene in primary cancers. Oncogene, 18, 3427–3430. [DOI] [PubMed] [Google Scholar]
- Milne G.T. and Weaver,D.T. (1993) Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev., 7, 1755–1765. [DOI] [PubMed] [Google Scholar]
- Mizuta R., LaSalle,J.M., Cheng,H.L., Shinohara,A., Ogawa,H., Copeland,N., Jenkins,N.A., Lalande,M. and Alt,F.W. (1997) RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc. Natl Acad. Sci. USA, 94, 6927–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C., Shinohara,A., Sonoda,E., Yamaguchi-Iwai,Y., Takata,M., Weichselbaum,R.R. and Takeda,S. (1999) The essential functions of human Rad51 are independent of ATP hydrolysis. Mol. Cell. Biol., 19, 6891–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen U.H., Bendixen,C., Sunjevaric,I. and Rothstein,R. (1996) DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA, 93, 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu,J.W., Koller,B.H. and Jasin,M. (1999) Brca1 controls homology-directed DNA repair. Mol. Cell, 4, 511–518. [DOI] [PubMed] [Google Scholar]
- Park M.S. (1995) Expression of human RAD52 confers resistance to ionizing radiation in mammalian cells. J. Biol. Chem., 270, 15467–15470. [DOI] [PubMed] [Google Scholar]
- Pierce A.J., Johnson,R.D., Thompson,L.H. and Jasin,M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev., 13, 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C.M. (1989) Helical RecA nucleoprotein filaments mediate homologous pairing and strand exchange. Biochim. Biophys. Acta, 1008, 131–145. [DOI] [PubMed] [Google Scholar]
- Radding C.M. (1991) Helical interactions in homologous pairing and strand exchange driven by RecA protein. J. Biol. Chem., 266, 5355–5358. [PubMed] [Google Scholar]
- Rattray A.J. and Symington,L.S. (1995) Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics, 139, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny Y., Rouillard,D., Chaput,B., Soussi,T. and Lopez,B.S. (1999) Mutant p53 proteins stimulate spontaneous and radiation-induced intrachromosomal homologous recombination independently of the alteration of the transactivation activity and of the G1 checkpoint. Oncogene, 18, 3553–3563. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schild D., Lio,Yc., Collins,D.W., Tsomondo,T. and Chen,D.J. (2000) Evidence for simultaneous protein interactions between human RAD51 paralogs. J. Biol. Chem., 275, 16443–16449. [DOI] [PubMed] [Google Scholar]
- Schoenmakers E.F., Huysmans,C. and Van de Ven,W.J. (1999) Allelic knockout of novel splice variants of human recombination repair gene RAD51B in t(12;14) uterine leiomyomas. Cancer Res., 59, 19–23. [PubMed] [Google Scholar]
- Scully R., Chen,J., Plug,A., Xiao,Y., Weaver,D., Feunteun,J., Ashley,T. and Livingston,D.M. (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell, 88, 265–275. [DOI] [PubMed] [Google Scholar]
- Shen Z., Cloud,K.G., Chen,D.J. and Park,M.S. (1996) Specific interactions between the human RAD51 and RAD52 proteins. J. Biol. Chem., 271, 148–152. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A. and Guarente,L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Buerstedde,J.M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchi-Iwai,Y. and Takeda,S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Morrison,C., Yamaguchi-Iwai,Y., Takata,M. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzbecher H.W., Donzelmann,B., Henning,W., Knippschild,U. and Buchhop,S. (1996) p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J., 15, 1992–2002. [PMC free article] [PubMed] [Google Scholar]
- Sun H., Treco,D., Schultes,N.P. and Szostak,J.W. (1989) Double-strand breaks at an initiation site for meiotic gene conversion. Nature, 338, 87–90. [DOI] [PubMed] [Google Scholar]
- Szostak J.W., Orr-Weaver,T.L., Rothstein,R.J. and Stahl,F.W. (1983) The double-strand-break repair model for recombination. Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- Thacker J. (1999) A surfeit of RAD51-like genes? Trends Genet., 15, 166–168. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T., Fujii,Y., Sakumi,K., Tominaga,Y., Nakao,K., Sekiguchi,M., Matsushiro,A., Yoshimura,Y. and MoritaT. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA, 93, 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyck E., Stasiak,A.Z., Stasiak,A. and West,S.C. (1999) Binding of double-strand breaks in DNA by human Rad52 protein [see comments]. Nature, 398, 728–731. [DOI] [PubMed] [Google Scholar]
- Vispé S., Cazaux,C., Lesca,C. and Defais,M. (1998) Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res., 26, 2859–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]