Abstract
Pyrosequencing is a method used to sequence DNA by detecting the pyrophosphate (PPi) group that is generated when a nucleotide is incorporated into the growing DNA strand in polymerase reaction. However, this method has an inherent difficulty in accurately deciphering the homopolymeric regions of the DNA templates. We report here the development of a method to solve this problem by using nucleotide reversible terminators. These nucleotide analogues are modified with a reversible chemical moiety capping the 3′-OH group to temporarily terminate the polymerase reaction. In this way, only one nucleotide is incorporated into the growing DNA strand even in homopolymeric regions. After detection of the PPi for sequence determination, the 3′-OH of the primer extension products is regenerated through different deprotection methods. Using an allyl or a 2-nitrobenzyl group as the reversible moiety to cap the 3′-OH of the four nucleotides, we have synthesized two sets of 3′-O-modified nucleotides, 3′-O-allyl-dNTPs and 3′-O-(2-nitrobenzyl)-dNTPs as reversible terminators for pyrosequencing. The capping moiety on the 3′-OH of the DNA extension product is efficiently removed after PPi detection by either a chemical method or photolysis. To sequence DNA, templates containing homopolymeric regions are immobilized on Sepharose beads, and then extension–signal detection–deprotection cycles are conducted by using the nucleotide reversible terminators on the DNA beads to unambiguously decipher the sequence of DNA templates. Our results establish that this reversible-terminator-pyrosequencing approach can be potentially developed into a powerful methodology to accurately determine DNA sequences.
Keywords: nucleotide reversible terminator, sequencing by synthesis
DNA sequencing is a fundamental tool for biological science. The completion of the Human Genome Project has set the stage for screening genetic mutations to identify disease genes on a genome-wide scale (1). Accurate high-throughput DNA sequencing methods are needed to explore the complete human genome sequence for applications in clinical medicine and health care. To overcome the limitations of the current electrophoresis-based sequencing technology (2–5), a variety of new DNA-sequencing methods have been investigated with an aim to eventually realize the goal of the $1,000 genome. Such approaches include sequencing by hybridization (6), mass spectrometry-based sequencing (7–9), sequence-specific detection of DNA using engineered nanopores (10), and sequencing by ligation (11). More recently, DNA sequencing by synthesis approaches such as pyrosequencing (12), sequencing of single DNA molecules (13, 14), and polymerase colonies (15) have been widely explored.
Pyrosequencing is a method to sequence DNA by detecting the pyrophosphate (PPi) that is generated when a nucleotide is incorporated into the growing DNA strand in polymerase reaction (12). In this approach, each of the four nucleotides is added sequentially with a mixture of enzymes and substrates in addition to the usual polymerase reaction components. If the added nucleotide is complementary with the first available base on the template, the nucleotide will be incorporated and a PPi will be released. The PPi is used by ATP sulfurylase to convert adenosine 5′-phosphosulfate to ATP, which provides the energy to the luciferase-mediated conversion of luciferin to oxyluciferin, which generates visible light. If the added nucleotide is not incorporated, no light will be produced and the nucleotide will simply be washed away or degraded by the enzyme apyrase. Pyrosequencing has been widely used in single nucleotide polymorphism detection and DNA methylation analysis (16, 17). More recently, this method was used in picoliter-sized reactors to produce the sequence of the known genome of Mycoplasma genitalium bacteria (18). However, the pyrosequencing method has an inherent problem in deciphering the number of bases in homopolymeric regions of DNA (12). The reason is that the light signal intensity is not exactly proportional to the amount of PPi released, especially when the homopolymeric region has more than five bases. Previously, we have reported the development of a general strategy to rationally design cleavable fluorescent nucleotide reversible terminators (NRTs) for four-color DNA sequencing by synthesis (19–23). In this approach, four nucleotides (A, C, G, and T) are modified as reversible terminators by attaching a cleavable fluorophore to the specific location of the base and capping the 3′-OH with a small chemically reversible moiety so that they are still recognized by DNA polymerase as substrates. DNA templates consisting of homopolymer regions were accurately sequenced by this approach (23). A recently developed sequencing-by-synthesis fluorescent DNA system based on a similar design of the cleavable fluorescent NRTs has already found wide applications in genome biology (24–26). Based on these successful results, we reasoned that we should be able to solve the homopolymer sequencing problem in conventional pyrosequencing by using four nucleotide analogues whose 3′-OH group is capped by a reversible moiety. We report here the design and synthesis of the 3′-O-allyl and 3′-O-(2-nitrobenzyl)-modified nucleotides and their successful application as reversible terminators for pyrosequencing to accurately decipher the homopolymeric regions of DNA.
Results and Discussion
Design and Synthesis of Cleavable NRTs for Pyrosequencing.
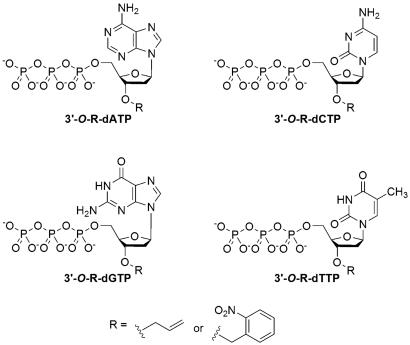
During the polymerase extension reaction, the 3′-OH group of the primer attacks the α-phosphate of the incoming nucleoside triphosphate to produce a DNA extension product, releasing a PPi molecule. Thus, when the NRTs, which have a reversible chemical moiety capping the 3′-OH group, are used to perform the polymerase reaction, the reaction will be temporarily terminated whenever a NRT is incorporated into the growing DNA strand. After the removal of the capping moiety, the polymerase reaction will resume. Based on this rationale, we synthesized and evaluated two sets of nucleotide analogues as NRTs for pyrosequencing: 3′-O-allyl-dNTPs and 3′-O-(2-nitrobenzyl)-dNTPs (Fig. 1). The allyl group can be efficiently removed by Pd-catalyzed deallylation, and the removal of the 2-nitrobenzyl moiety is readily accomplished by laser irradiation at 355 nm. The design and synthesis of the 3′-O-allyl-dNTPs has been described previously (23).
Fig. 1.
Structures of NRTs 3′-O-allyl-dNTP and 3′-O-(2-nitrobenzyl)-dNTP.
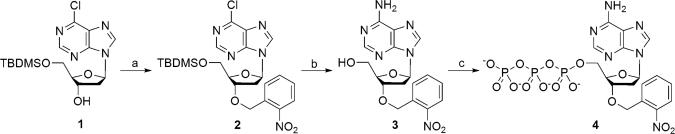
It is particularly challenging to synthesize 3′-O-(2-nitrobenzyl)-dNTPs because the nucleophilic nitrogen on the base preferentially reacts with the 2-nitrobenzyl group. Using a previously reported method (27) for the synthesis of a 3′-O-(2-nitrobenzyl)-dATP actually led to the final nucleotide analogue with the 2-nitrobenzyl group attached to the 6-amino group of the purine base (28). We have developed a selective protection strategy for the synthesis of four 3′-O-(2-nitrobenzyl)-dNTPs, and the detailed procedures are described in supporting information (SI) Appendix. The synthesis of 3′-O-(2-nitrobenzyl)-dATP is shown in Fig. 2 as an example. Treatment of 9-[β-d-5′-O-(tert-butyldimethylsilyl)-2′-deoxyribofuranosyl]-6-chloropurine 1, in which both the sugar and base were modified to allow the site-specific introduction of the 2-nitrobenzyl group to the 3′-oxygen, with 2-nitrobenzyl bromide under basic conditions furnished 2-nitrobenzylated compound 9-[β-d-5′-O-(tert-butyldimethylsilyl)-3′-O-(2-nitrobenzyl)-2′-deoxyribofuranosyl]-6-chloropurine 2, which was desilylated and converted to 2-deoxyadenosine derivative 3′-O-(2-nitrobenzyl)-2′-deoxyadenosine 3 in a one-pot reaction. The precursor 3 was then transformed to the target molecule 3′-O-(2-nitrobenzyl)-dATP 4 with established triphosphorylation procedures (23, 29, 30).
Fig. 2.
Synthesis of 3′-O-(2-nitrobenzyl)-dATP. (Step a) 2-nitrobenzyl bromide, tetrabutylammonium bromide, NaOH, in CH2Cl2 at room temperature for 1 h to produce compound 2 with a 95% yield. (Step b) Tetrabutylammonium fluoride in THF at room temperature for 1 h; methanolic ammonia and dioxane at 85–90°C for 12 h to produce compound 3 with a 56% yield. (Step c) POCl3, PO(OMe)3 at 0°C for 2 h; (Bu3NH)4P2O7, Bu3N, triethylammonium bicarbonate, and NH4OH at room temperature for 1.5 h to produce compound 4 with a 30% yield.
Polymerase Extension Using 3′-O-modified Nucleotides and Characterization by MALDI-TOF MS.
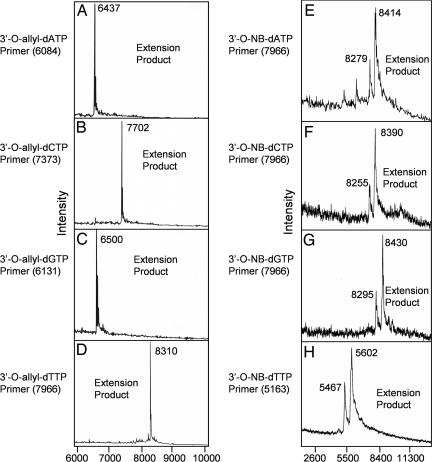
3′-O-modified nucleotides pose a great challenge for incorporation by natural polymerase, especially when the 3′-O-labeling group is a bulky one (31, 32). To verify that the NRTs can be recognized by polymerase as substrates in a polymerase reaction, we performed extension reactions with four different primers corresponding to different regions of a DNA template whose next complementary base was either A, C, G, or T. A 9°N polymerase (exo-)A485L/Y409V, which has been shown previously to incorporate the 3′-O-modified nucleotides (21, 23), was used in the polymerase extension reaction. After the reaction, the eight different primer extension products [four for 3′-O-allyl-dNTPs and four for 3′-O-(2-nitrobenzyl)-dNTPs] were analyzed by MALDI-TOF MS, and the results are shown in Fig. 3. Single clear mass peaks at 6,437, 7,702, 6,500, and 8,310 (m/z) for each primer extension product was produced by using 3′-O-allyl-dNTPs with no left-over primer peak (Fig. 3 A–D). Similarly, the 3′-O-(2-nitrobenzyl)-dNTPs also produced complete primer extension DNA products at 8,414, 8,390, 8,430, and 5,602 (m/z) (Fig. 3 E–H). The small peaks at 8,279, 8,255, 8,295, and 5,467 (m/z) in the mass spectra for the 3′-O-(2-nitrobenzyl)-dNTP extension products correspond to the photocleavage products that were generated by the partial photocleavage of the DNA extension products induced by the nitrogen laser (337 nm) used for ionization of the analyte in MALDI-TOF MS. These results indicate that the primers were quantitatively extended by the 3′-O-modified-dNTPs in polymerase reaction and that the modified nucleotides are excellent substrates for the 9°N polymerase.
Fig. 3.
MALDI-TOF MS spectra of primer extension products with 3′-O-allyl-dNTPs (A–D) and 3′-O-(2-nitrobenzyl)-dNTPs (E–H). All eight 3′-O-modified nucleotides are quantitatively incorporated into the primers with high efficiency in the polymerase reaction, which indicates that the modified nucleotides are good substrates for the polymerase. The small peak near the 3′-O-(2-nitrobenzyl)-dNTP extension product corresponds to the photocleaved product generated during the laser desorption and ionization process used in MALDI-TOF MS.
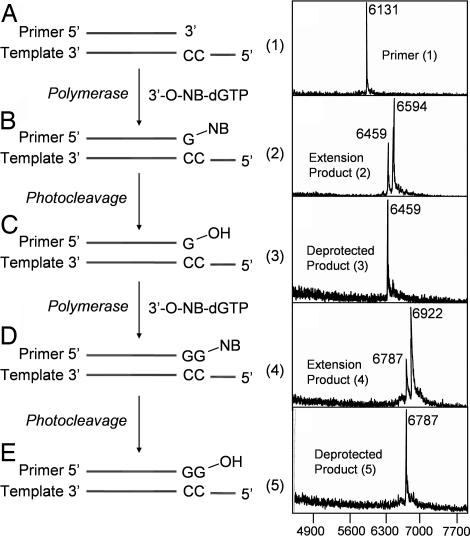
To further verify the utility of the NRTs in determining the homopolymeric regions of DNA sequences, we performed a continuous polymerase extension reaction in solution. This procedure allows the isolation of the DNA product at each step for detailed molecular characterization by MALDI-TOF MS. First, a polymerase extension reaction using 3′-O-(2-nitrobenzyl)-dGTP as a reversible terminator along with a primer and synthetic 100-mer DNA template corresponding to a portion of exon 7 in the human p53 gene was performed to yield a single-base extension product (product 2) (Fig. 4B Left ). After the reaction, a small portion of the extension product was characterized by MALDI-TOF MS. The rest of the product was irradiated with a laser at 355 nm for 30 s to cleave the 3′-O-(2-nitrobenzyl) group from the DNA to yield photocleaved product (product 3) (Fig. 4C Left), which was characterized by MALDI-TOF MS. The photocleaved DNA product (product 3) with a free 3′-OH group regenerated was then used as a primer for the next nucleotide extension reaction. Fig. 4 A Right–E Right shows the sequential mass spectrum at each step of continuous DNA extension reaction using 3′-O-(2-nitrobenzyl)-dGTP as a reversible terminator. The primer alone produces a peak at 6,131 (m/z) (Fig. 4A). The mass peak at 6,594 (m/z) in Fig. 4B corresponds to the first extension product with a single modified nucleotide G incorporated in this homopolymeric region. The small peak at 6,459 (m/z) in Fig. 4B corresponds to the photocleavage product that was generated by the nitrogen laser (337 nm) used for ionization of the analyte in MALDI-TOF MS. Fig. 4C shows the photocleavage result after irradiation of the extension product (product 2) at 355 nm. It can be seen from the data that the peak at 6,594 (m/z) has completely vanished, and only a single peak corresponding to the DNA product (product 3) remains at 6,459 (m/z), which indicates that the 2-nitrobenzyl moiety was efficiently removed to regenerate the 3′-OH group. Fig. 4D shows the MALDI-TOF MS data for the extension product obtained by using the photocleaved DNA product (compound 3) as a primer to incorporate another 3′-O-(2-nitrobenzyl)-dGTP. A dominant peak is seen at 6,922 (m/z) corresponding to the extension product (product 4). The small peak at 6,787 (m/z) corresponds to the photocleavage product that was generated by the nitrogen laser (337 nm) used for ionization of the analyte in MALDI-TOF MS. Upon further photolysis at 355 nm, the 2-nitrobenzyl moiety was removed to yield DNA product (product 5) at 6,787 (m/z) with a free 3′-OH group (Fig. 4E). Similar data were obtained for 3′-O-(2-nitrobenzyl)-dTTP (SI Fig. 8). The other two nucleotides, 3′-O-(2-nitrobenzyl)-dATP, and 3′-O-(2-nitrobenzyl)-dCTP also were verified to be excellent reversible terminators for the 9°N polymerase.
Fig. 4.
The polymerase extension scheme using 3′-O-(2-nitrobenzyl)-dGTP (A Left–E Left) and MALDI-TOF MS spectra of the two consecutive extension products and their photocleavage products (A Right–E Right). (A) Primer for the polymerase extension reaction. (B) Primer extended with 3′-O-(2-nitrobenzyl)-dGTP to yield DNA extension product 2. (C) Product 2 photocleaved to yield photocleavage product 3. (D) Product 3 extended with another 3′-O-(2-nitrobenzyl)-dGTP to yield product 4. (E) Product 4 photocleaved to yield photocleavage product 5. After 30 s of irradiation with a laser at 355 nm, photocleavage is complete with all of the 3′-O-(2-nitrobenzyl)-group cleaved from the DNA extension products.
3′-O-Modified dATP Is Not a Substrate of Luciferase.
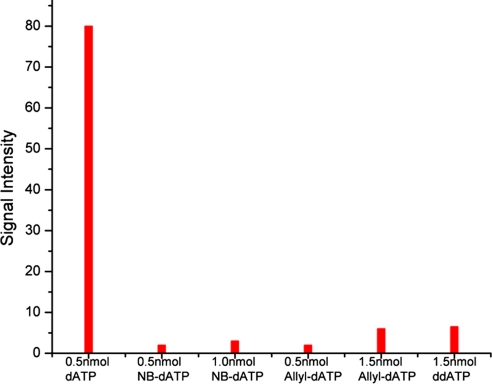
In pyrosequencing, luciferase converts luciferin to oxyluciferin by using the energy provided by ATP, yielding a chemiluminescence light signal. However, the natural nucleotide dATP also is a substrate of luciferase, which can produce a false positive signal to seriously interfere with the pyrosequencing result. To solve this problem, a sulfur-modified nucleotide, α-S-dATP, which is not a substrate for luciferase, is used instead of the natural dATP in conventional pyrosequencing (33). To our delight, the 3′-O-modified-dATPs [3′-O-allyl-dATP and 3′-O-(2-nitrobenzyl)-dATP] were shown not to be substrates of luciferase as indicated by the data in Fig. 5. 3′-O-modified-dATP and dATP were separately added to the luciferase and luciferin mixtures and the corresponding light intensities were measured and compared. dATP (0.5 nmol) produced a light signal intensity of 80, whereas 0.5 nmol and 1.5 nmol of 3′-O-modified-dATP only led to light intensities near background level. These results confirmed that 3′-O-modified-dATP is not a substrate of luciferase. Fig. 5 also shows that ddATP is not a substrate to luciferase. These results indicate that the 3′-OH group may play a significant role in luciferase-catalyzed reaction. This hydroxyl group may interact with the active catalytic site of the luciferase. This interaction is interrupted when the 3′-OH group is modified with an allyl group or a 2-nitrobenzyl group (or without a 3′-hydroxyl group as in ddATP), thereby preventing luciferase from using 3′-O-modified-dATP and ddATP as a substrate. Thus, 3′-O-modified-dATP can be directly used in pyrosequencing without any further modification.
Fig. 5.
Signal intensity of luciferase catalyzed reactions using 0.5 nmol of dATP, 0.5 nmol of 3′-O-(2-nitrobenzyl)-dATP, 1.0 nmol of 3′-O-(2-nitrobenzyl)-dATP, 0.5 nmol of 3′-O-allyl-dATP, 1.5 nmol of 3′-O-allyl-dATP, and 1.5 nmol of ddATP. The results show that 3′-O-(2-nitrobenzyl)-dATP and 3′-O-allyl-dATP are not substrates of luciferase (NB, 2-nitrobenzyl).
Pyrosequencing with 3′-O-Modified NRTs.
To verify that the NRTs can be successfully used in pyrosequencing, we carried out a sequencing reaction on a self-priming DNA template, which contained multiple homopolymeric regions, immobilized on Sepharose beads (SI Fig. 9). The pyrosequencing reaction was initiated by extending the DNA template using a polymerase extension reaction mixture containing the NRTs. The extension of the primer by only the complementary NRT was confirmed by subsequent enzymatic cascade reactions to convert the released PPi into a light signal. For the 3′-O-allyl-dNTPs, after detection of the light signal, the DNA beads were immersed in a Pd deallylation solution and incubated for 2 min to cleave the 3′-O-allyl group to regenerate a free 3′-OH for further extension. In the case of 3′-O-(2-nitrobenzyl)-dNTPs, after NRT incorporation, the DNA beads were irradiated with a laser at 355 nm to remove the 2-nitrobenzyl group for further extension. After washing the beads, the next extension cycle was initiated. Extension–signal detection–deprotection cycles were performed multiple times to decipher unambiguously the homopolymeric sequences in the DNA template.
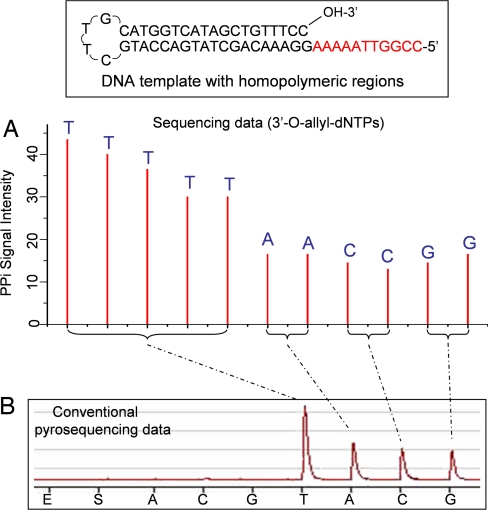
The pyrosequencing data generated by 3′-O-allyl-dNTPs are shown in Fig. 6A. The 11 bases in the homopolymeric regions (five T, two A, two C, and two G bases) are clearly identified, whereas the pyrosequencing data obtained by using natural nucleotides show a single large peak corresponding to a stretch of Ts and three smaller peaks corresponding to stretches of A, C, and G bases (Fig. 6B). However, it is very difficult to identify the exact sequence from this conventional pyrosequencing data.
Fig. 6.
Comparison of reversible terminator-pyrosequencing using 3′-O-allyl-dNTPs with conventional pyrosequencing using natural nucleotides. (A) The self-priming DNA template with stretches of homopolymeric regions (five A, two T, two G, and two C bases) was sequenced by using 3′-O-allyl-dNTPs. The homopolymeric regions are clearly identified, with each peak corresponding to the identity of each base in the DNA template. (B) Pyrosequencing data using natural nucleotides. The homopolymeric regions produced one large peak corresponding to the stretch of T bases and three smaller peaks for stretches of A, C, and G bases. However, it is very difficult to decipher the exact sequence from the data.
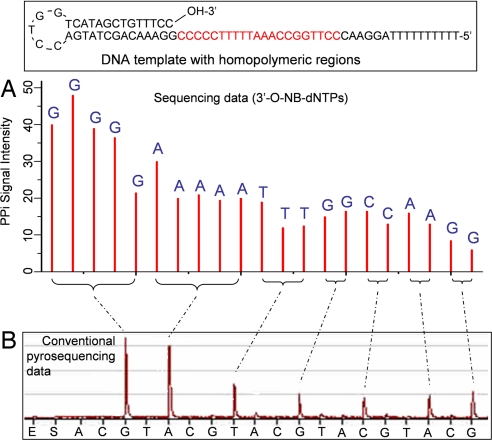
The pyrosequencing results using 3′-O-(2-nitrobenzyl)-dNTPs are shown in Fig. 7A. Twenty-one bases in the homopolymeric regions (five G, five A, three T, two G, two C, two A, and two G bases) are clearly identified, whereas the pyrosequencing data obtained by using natural nucleotides shows two large peaks corresponding to stretches of G and A bases and five smaller peaks corresponding to stretches of T, G, C, A and G bases (Fig. 7B), leading to ambiguity to identify the sequence. To further verify the utility of the reversible terminator-pyrosequencing method, we used 3′-O-(2-nitrobenzyl)-dNTPs to sequence a PCR DNA template produced by amplification on Sepharose beads to unambiguously decipher 11 bases in the DNA templates containing homopolymeric sequences (SI Appendix and SI Fig. 10).
Fig. 7.
Comparison of reversible terminator-pyrosequencing using 3′-O-(2-nitrobenzyl)-dNTPs with conventional pyrosequencing using natural nucleotides (NB, 2-nitrobenzyl). (A) The self-priming DNA template with stretches of homopolymeric regions was sequenced by using 3′-O-(2-nitrobenzyl)-dNTPs. The homopolymeric regions are clearly identified, with each peak corresponding to the identity of each base in the DNA template. (B) Pyrosequencing data using natural nucleotides. The homopolymeric regions produced two large peaks corresponding to the stretches of G and A bases and five smaller peaks corresponding to stretches of T, G, C, A, and G bases. However, it is very difficult to decipher the exact sequence from the data.
Conclusion
We have developed two sets of NRTs, 3′-O-allyl-dNTP and 3′-O-(2-nitrobenzyl)-dNTP, for pyrosequencing, which are able to accurately decipher the homopolymeric sequences in DNA templates. The reversible terminators were efficiently incorporated, and they terminated the polymerase reactions, and the released PPi for each extension was detected with a standard luciferase assay. We have generated preliminary feasibility sequencing data of 11 bases with 3′-O-allyl-dNTPs and 21 bases with 3′-O-(2-nitrobenzyl)-dNTPs on DNA templates consisting of multiple homopolymer regions. Longer read length should be possible with further optimization in nucleotide incorporation efficiency and deprotection efficiency coupled with automation. Also, other alternative reversible chemical groups can be explored for further optimization of the NRTs for pyrosequencing. In addition to solving the homopolymer issues in conventional pyrosequencing, the other advantage of using the NRTs in extension–signal detection–deprotection cycles is that higher efficiency can be achieved with multiple extensions or deprotections without any dephasing in the sequence determination or a reduction in the sequencing accuracy. Therefore, in principle, one can achieve >99% efficiency in each cycle to reach read lengths of at least several hundred. The signal reduction in our preliminary pyrosequencing data generated with the NRTs is mainly due to the loss of DNA beads during each washing step because the reaction was performed manually. Therefore, longer read lengths can be achieved when using single DNA-bead extension and automated washing systems, such as the 454 genome sequencer (18). It is well established that PCR templates can be generated on millions of beads through emulsion PCR (18, 34). Thus, future implementation of the reversible-terminator pyrosequencing on a high-density bead array platform will provide a high-throughput and accurate DNA sequencing system with wide applications in genome biology and biomedical research.
Materials and Methods
Synthesis of 3′-O-Allyl-dNTPs and 3′-O-(2-Nitrobenzyl)-dNTPs.
3′-O-allyl-dNTPs were synthesized according to the literature (23), and the synthesis of 3′-O-(2-nitrobenzyl)-dNTP is described in SI Appendix. An enzymatic method was used to yield ultrapure 3′-O-modified nucleotide analogues based on the literature (35) (also see SI Appendix).
Incorporation of 3′-O-Modified NRTs in Solution and Characterization by MALDI-TOF MS.
Each polymerase extension reaction solution consists of 40 pmol of templates, 40 pmol of primers (the template and primer sequences are described in SI Table 1), 100 pmol of NRTs, 2 μl of 10× Thermopol II reaction buffer (New England Biolabs, Ipswich, MA), 2 μl of 20 mM MnCl2, and 2 μl (4 units) of 9°N polymerase (exo-)A485L/Y409V in a total volume of 20 μl. After an initial incubation at 95°C for 5 min and 4°C for 5 min, the reaction was performed at 95°C for 15 seconds, 55°C for 15 seconds, and 65°C for 1 min for 20 cycles. The resulting DNA products were purified for MALDI-TOF MS analysis by using a previously reported procedure (23). We also characterized 3′-O-(2-nitrobenzyl)-dGTP by performing a continuous DNA extension reaction using a primer (5′-GTTGATGTACACATTGTCAA-3′) and a synthetic DNA template (SI Table 1). The detailed procedure is described in SI Appendix. The other 3′-O-(2-nitrobenzyl)-dNTPs were similarly characterized.
Pyrosequencing Using the NRTs.
Each extension reaction consisted of Sepharose bead-immobilized DNA (the procedure to prepare the DNA beads is described in SI Appendix), 200 pmol of NRTs, 1.2 μl of 50 mM MnCl2, 1 μl (2 units) of 9°N polymerase (exo-)A485L/Y409V, and 20 μl of annealing buffer (20 mM Tris-acetate/5 mM magnesium acetate, pH 7.6). Extension was conducted in a thermal cycler and incubated at 65°C for 20 min with occasional stirring to prevent the beads from settling. After the polymerase reaction, the beads were pelleted by centrifugation for 20 s, and the supernatant was carefully removed. The beads were washed with 30 μl of annealing buffer, and the PPi of the combined supernatant was detected on a 96PSQ Pyrosequencer (Biotage, Uppsala, Sweden) for sequence determination (33). After detection of the signal, the beads were washed three times with 180 μl of deionized water. For 3′-O-allyl-dNTP extensions, deallylation was conducted under aqueous-Pd-catalyzed conditions (23). After deallylation, the beads were washed three times with 180 μl of 1 M Tris-acetate buffer (pH 7.7) and three times with 180 μl of annealing buffer and the next extension–signal detection–deprotection cycle was initiated. For 3′-O-(2-nitrobenzyl)-dNTP extensions, extended DNA beads were suspended in 1 ml of annealing buffer in a cuvette with stirring and irradiated with a laser at 355 nm (3 W/cm2) for 1 min. After photocleavage, the beads were washed two times with annealing buffer for the continuation of the subsequent extension reactions. Conventional pyrosequencing data shown in Figs. 6B and 7B were generated by using the same instrument in parallel to compare the data with those of pyrosequencing by using the NRTs.
Supplementary Material
Acknowledgments
We thank Dr. Steffen Jockusch, Dr. James J. Russo, and Mr. Liyong Deng for generous discussions and technical support. This work was supported by National Institutes of Health Grants P50 HG002806, R01 HG003582, and R21HG004404 and by the Packard Fellowship for Science and Engineering.
Abbreviations
- NRT
nucleotide reversible terminator
- PPi
pyrophosphate.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707495104/DC1.
References
- 1.Collins FS, Green ED, Guttmacher AE, Guyer MS. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F, Nicklen S, Coulson AR. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maxam AM, Gilbert W. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SB, Hood LE. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 5.Ju J, Ruan C, Fuller CW, Glazer AN, Mathies RA. Proc Natl Acad Sci USA. 1995;92:4347–4351. doi: 10.1073/pnas.92.10.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drmanac S, Kita D, Labat I, Hauser B, Schmidt C, Burczak JD, Drmanac R. Nat Biotechnol. 1998;16:54–58. doi: 10.1038/nbt0198-54. [DOI] [PubMed] [Google Scholar]
- 7.Fu DJ, Tang K, Braun A, Reuter D, Darnhofer-Demar B, Little DP, O'Donnell MJ, Cantor CR, Koster H. Nat Biotechnol. 1998;16:381–384. doi: 10.1038/nbt0498-381. [DOI] [PubMed] [Google Scholar]
- 8.Roskey MT, Juhasz P, Smirnov IP, Takach EJ, Martin SA, Haff LA. Proc Natl Acad Sci USA. 1996;93:4724–4729. doi: 10.1073/pnas.93.10.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards JR, Itagaki Y, Ju J. Nucleic Acids Res. 2001;29:E104. doi: 10.1093/nar/29.21.e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 12.Ronaghi M, Uhlen M, Nyrén P. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 13.Braslavsky I, Hebert B, Kartalov E, Quake SR. Proc Natl Acad Sci USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenleaf WJ, Block SM. Science. 2006;313:801. doi: 10.1126/science.1130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitra RD, Shendure J, Olejnik J, Olejnik EK, Church GM. Anal Biochem. 2003;320:55–65. doi: 10.1016/s0003-2697(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 16.Nordstrom T, Ronaghi M, Forsberg L, de Faire U, Morgenstern R, Nyren P. Biotechnol Appl Biochem. 2000;31:107–112. doi: 10.1042/ba19990104. [DOI] [PubMed] [Google Scholar]
- 17.Tost J, Dunker J, Glynne GY. BioTechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 18.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju J, Li Z, Edwards J, Itagaki Y. US Patent 6. 2003;664:079. [Google Scholar]
- 20.Li Z, Bai X, Ruparel H, Kim S, Turro NJ, Ju J. Proc Natl Acad Sci USA. 2003;100:414–419. doi: 10.1073/pnas.242729199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruparel H, Bi L, Li Z, Bai X, Kim DH, Turro NJ, Ju J. Proc Natl Acad Sci USA. 2005;102:5932–5937. doi: 10.1073/pnas.0501962102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo TS, Bai X, Kim DH, Meng Q, Shi S, Ruparel H, Li Z, Turro NJ, Ju J. Proc Natl Acad Sci USA. 2005;102:5926–5931. doi: 10.1073/pnas.0501965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ju J, Kim DH, Bi L, Meng Q, Bai X, Li Z, Li X, Marma MS, Shi S, Wu J, et al. Proc Natl Acad Sci USA. 2006;103:19635–19640. doi: 10.1073/pnas.0609513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson DS, Mortazavi A, Myers RM, Wold B. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 26.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Metzker ML, Raghavachari R, Richards S, Jacutin SE, Civitello A, Burgess K, Gibbs RA. Nucleic Acids Res. 1994;22:4259–4267. doi: 10.1093/nar/22.20.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welch MB, Martinez CI, Zhang AZ, Jin S, Gibbs R, Burgess K. Chem Eur J. 2005;11:7145. [Google Scholar]
- 29.Kawate T, Allerson CR, Wolfe JL. Org Lett. 2005;7:3865–3868. doi: 10.1021/ol051144r. [DOI] [PubMed] [Google Scholar]
- 30.Ludwig J, Eckstein F. J Org Chem. 1989;54:631–635. [Google Scholar]
- 31.Welch MB, Martinez CI, Zhang AJ, Jin S, Gibbs R, Burgess K. Chem Eur J. 1999;5:951–960. [Google Scholar]
- 32.Lu G, Burgess K. Bioorg Med Chem Lett. 2006;16:3902–3905. doi: 10.1016/j.bmcl.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 33.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 34.Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 35.Metzker ML, Raghavachari R, Burgess K, Gibbs RA. BioTechniques. 1998;25:814–817. doi: 10.2144/98255st01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.