Abstract
Polyphosphate kinase 1 (PPK1), the principal enzyme responsible for reversible synthesis of polyphosphate (poly P) from the terminal phosphate of ATP, is highly conserved in bacteria and archaea. Dictyostelium discoideum, a social slime mold, is one of a few eukaryotes known to possess a PPK1 homolog (DdPPK1). Compared with PPK1 of Escherichia coli, DdPPK1 contains the conserved residues for ATP binding and autophosphorylation, but has an N-terminal extension of 370 aa, lacking homology with any known protein. Polyphosphate or ATP promote oligomerization of the enzyme in vitro. The DdPPK1 products are heterogeneous in chain length and shorter than those of E. coli. The unique DdPPK1 N-terminal domain was shown to be necessary for its enzymatic activity, cellular localization, and physiological functions. Mutants of DdPPK1, as previously reported, are defective in development, sporulation, and predation, and as shown here, in late stages of cytokinesis and cell division.
Keywords: inorganic polyphosphate, cell division, multinucleated cells
Inorganic polyphosphate (poly P), a linear chain of tens or hundreds of orthophosphate (Pi) residues linked by high-energy phosphoanhydride bonds, is ubiquitous in nature. It is a dynamic molecule in all organisms (archaea, bacteria, fungi, plants, insects, and mammals) and may attain a level of 20% of the cell dry weight in Saccharomyces cerevisiae (1). Conserved and versatile, poly P plays a variety of essential roles. Among them are survival in the face of stress and stringencies (2, 3), bacterial motility (4–6), biofilm formation and virulence (7), predation, and the developmental stages of social bacteria and slime molds (8, 9). Poly P is also involved in blood coagulation (10) and mammary cancer cell proliferation (11). Poly P is a part of a polyhydroxybutyrate–Ca2+ complex responsible for the uptake of DNA in Escherichia coli and ion transport in human mitochondria (12, 13).
Among the growing list of enzymes that make and hydrolyze poly P, polyphosphate kinase 1 (PPK1) is the most widely conserved (14). PPK1 homologs have been found in >100 prokaryotic species, including 20 major pathogens, and to date, only in a few eukaryotes, including the PPK1 homolog of the social slime mold Dictyostelium discoideum (DdPPK1) (15, 16). The deduced amino acid sequence of DdPPK1 shares 30% identity and 51% similarity with E. coli PPK1 (EcPPK1). However, DdPPK1 contains 1,050 amino acid residues as compared with only 688 residues in EcPPK1; the 370-aa N-terminal domain, occupying one-third of DdPPK1 in length, shows no homology to EcPPK1 or any other protein in the GenBank database.
The crystal structure of EcPPK1 (17) revealed an ATP-binding pocket located in a highly conserved structural tunnel. Among the 11 conserved residues essential for a presumed poly P tunnel, 10 are present in DdPPK1; 14 of 16 residues in the ATP binding pocket are also located in DdPPK1 [supporting information (SI) Fig. 8]. Studies with EcPPK1 indicated that two histidine residues, H435 and H454, are the probable autophosphorylation sites involved in poly P synthesis (18). However, recent studies implicated only H435 (17). The two histidine residues (H435, critical for autophosphorylation, and H592, required as a general acid catalyst in EcPPK1) and four other highly conserved residues (corresponding to D470, E623, S610, and T458 in EcPPK1) are also present in DdPPK1. Thus, DdPPK1 may share similar structural features with EcPPK1 in the unique ATP binding pocket and poly P tunnel.
Mutants deficient in DdPPK1 (herein after referred to as DdPPK1 mutant) have a reduced poly P level, and are defective in fruiting body development, sporulation, and phagocytosis (8). Here, we report the enzymatic features and the cellular localization of the DdPPK1 and the essential feature of its unique N-terminal domain in enzymatic activity, protein sorting, and cellular functions. Moreover, we also observed the deficiency of the mutant in cytokinesis, a fundamental process in cell division.
Results
DdPPK1 Activity.
A PPK1 activity of <200 units per mg of protein was detected in lysates of WT D. discoideum under standard EcPPK1 reaction conditions. The effect of reaction temperature on enzyme activity was tested between 15°C and 40°C, and the maximum activity was observed at 37°C. The specific activity was increased nearly 10-fold to the level of ≈2,000 units per mg of protein by adding poly P (type 15 or 75, 0.5–5 mM in Pi residues) and by optimizing salt concentration and pH (see Materials and Methods). When grown in HL5 medium, the activity peaked in log phase (1–2 × 106 cells per ml) and declined as cells entered stationary phase (SI Fig. 9).
Expression and Purification of DdPPK1.
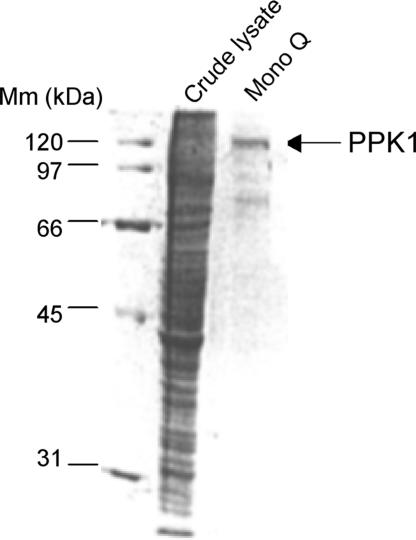
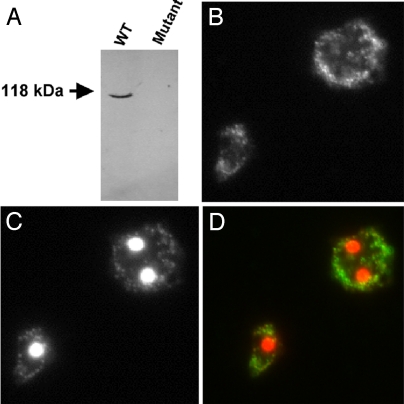
Ddppk1 was cloned into a D. discoideum expression vector pTX-GFP to yield pTX-Ddppk1 (8), in which Ddppk1 was overexpressed under the strong promoter Pact15. WT cells containing pTX-Ddppk1 have DdPPK1 activities of ≈10,000 units per mg of protein in crude lysates, at least 5-fold higher than WT cells. DdPPK1 was purified from the overexpressing cells by sequential chromatography on Heparin, DEAE, and Mono Q columns (Table 1). DdPPK1 was purified ≈500-fold compared with the enzyme in the WT; the specific activity of the purified protein was ≈5 × 105 units per mg of protein, a value near that of the recombinant EcPPK1 (2 × 106 units per mg of protein). The purified DdPPK1 has an approximate molecular mass of 118 kDa on SDS/PAGE (Fig. 1), indicating that DdPPK1 does not undergo major processing in vivo. The purified preparation did not contain a protein corresponding to DdPPK2 or a similar activity (19).
Table 1.
Purification of DdPPK1
| Sample | Specific activity, Units/mg × 104 | Total activity*, units × 105 | Purification, fold | Recovery, % |
|---|---|---|---|---|
| WT cell lysate | 0.1 | |||
| Overproducer | 1.0 | 17.7 | 10 | |
| Heparin | 15 | 6.7 | 150 | 38 |
| FT of DEAE | 17 | 5.4 | 170 | 30 |
| Mono Q column | 48 | 1.5 | 480 | 8.5 |
*One unit of enzyme is defined as the amount incorporating 1 pmol of phosphate into poly P per min at 37°C.
Fig. 1.
SDS/PAGE (12%) analysis of purified DdPPK1. Samples were precipitated with 0.015% deoxycholate/5% trichloroacetic acid. Proteins were visualized by Coomassie blue staining. Crude lysate (≈50 μg) and purified fraction (≈3 μg) (Mono Q column) were applied as indicated. Mm, protein molecular mass markers (Bio-Rad).
Oligomerization of DdPPK1.
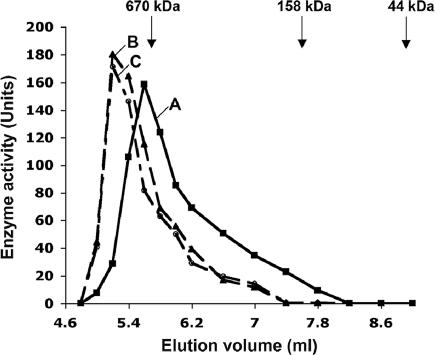
The molecular mass of native DdPPK1 was measured by size-exclusion chromatography on a HPLC system. On the basis of the elution pattern of DdPPK1 with Na2SO4 (150 mM, pH 7.4), the size of the protein was estimated to be ≈720 kDa, presumably a hexamer of 118-kDa subunit (Fig. 2). In the presence of poly P (type 15, 10 mM in Pi residues) or ATP (2 mM), DdPPK1 was eluted in the void volume (5.2 ml) of the column, suggesting a molecular mass of >1,028 kDa under these conditions. DdPPK1 may thus form higher oligomeric structures in the presence of either poly P or ATP.
Fig. 2.
Size-exclusion chromatography of DdPPK1. Lysates of D. discoideum DdPPK1 overexpressing cells were prepared and incubated in elution buffer only (A), elution buffer with 10 mM poly P (type 15) (B), or elution buffer with 2 mM ATP (C). After incubating at 0°C for 30 min, 200-ml lysates were applied onto a Tosoh G3000SWXL HPLC column and eluted with the same incubation buffer. Enzyme activity was measured in each fraction. Molecular mass standards were thyroglobulin (670 kDa), bovine gamma globulin (158 kDa), and chicken ovalbumin (44 kDa).
Products of DdPPK1.
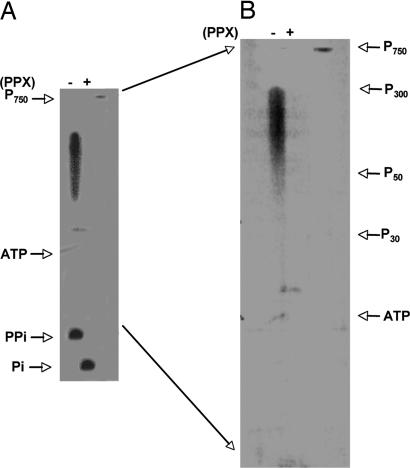
As with other PPK1s, the poly P synthesizing activity of DdPPK1 was processive, but the products were more heterogeneous and shorter in chain length (Fig. 3). Unlike EcPPK1, which produced a monodisperse long-chain poly P of 700–800 residues, DdPPK1 produced poly P with broad-range chain lengths of 50–300 Pi residues, among the shortest produced by any known PPK1s so far. The poly P products were completely hydrolyzed by an exopolyphosphatase (Fig. 3).
Fig. 3.
Poly P products of DdPPK1. Poly P synthesis with 40 ng of DdPPK1 was carried out with [γ-32P]ATP under the optimal conditions for 30 min. Products were purified, treated with exopolyphosphatase (PPX) (as described in Materials and Methods) and separated by 20% PAGE containing 7 M urea, and visualized by PhosphorImager (Molecular Dynamics). P750 is a purified product of EcPPK1, which has a chain length of ≈750 residues. P300, P50, and P30 indicate the position of poly P with different chain lengths. PPi, pyrophosphate. (A) Poly P products before (−) and after (+) PPX-treatment. (B) The region between poly P750 and PPi from gel A is magnified to highlight the chain lengths of poly P.
Other Characteristics of DdPPK1.
Poly P is known to stimulate the activities of Pseudomonas aeruginosa PPK2 (16) and Myxococcus xanthus PPK1 (9). A nearly 10-fold stimulation was observed for DdPPK1 in crude lysates, but stimulation was <2-fold with the purified enzyme when poly P was added to the reaction mixture (SI Fig. 10). In crude lysates, the added poly P may act to protect the 32P-poly P product from degradation.
D. discoideum also has an actin-related poly P synthesizing complex (DdPPK2) that is KCl dependent (19). In contrast, KCl inhibited DdPPK1 activity (SI Fig. 11A). At a concentration of 200 mM KCl, the inhibition was 80%, and at 500 mM, the inhibition was almost complete.
Phalloidin, a heptapeptide toxin from the mushroom Amanita phalloides, which binds tightly and specifically to polymerized actin (20), inhibited poly P synthesis by DdPPK2 (19). Assuming that DdPPK2 was a tetramer, complete inhibition was seen at a molar ratio of phalloidin to the enzyme between 1:1 and 1:2. However, DdPPK1 activity was inhibited only by 25% at 2 nM phalloidin and ≈80% at 10 nM (SI Fig. 11B). The molar ratio of phalloidin to DdPPK1 is 10:15 for complete inhibition of DdPPK1 activity (SI Fig. 11C). Similar ratios of phalloidin and PPK1 were observed for inhibition of PPK1 from E. coli, P. aeruginosa, and Bacillus cereus (M.R.G.-G. and X.S., unpublished data).
Localization of DdPPK1.
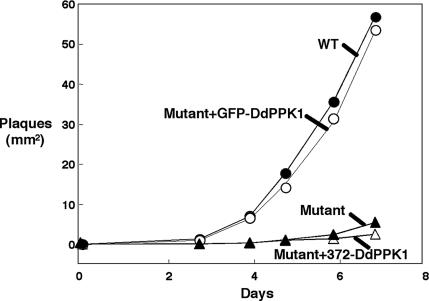
The localization of DdPPK1 was detected by using a GFP-DdPPK1 fusion protein and by immunofluorescence methods. A GFP-DdPPK1 fusion protein was both enzymatically and functionally active. The mutant carrying pTX-gfp-Ddppk1 expressed PPK1 activity up to levels of ≈5,000 units/mg in a crude lysate (Table 2) and was similar to WT in plaque-forming activity on a Klebsiella aerogenes lawn (Fig. 4).
Table 2.
PPK activities of different D. discoideum cells
| D. discoideum strains | Specific activity of crude lysate, units/mg |
|---|---|
| WT | 1,435 |
| Mutant (Ddppk1-) | 188 |
| Mutant (pTX-gfp) | 156 |
| Mutant (pTX-Ddppk1) | 11,027 |
| Mutant (pTX-gfp-Ddppk1) | 5,340 |
| Mutant (pTX-372-Ddppk1) | 284 |
| Mutant (pTX-gfp-373-Ddppk1) | 228 |
Note that pTX-gfp is the vector; pTX-Ddppk1 and pTX-gfp-Ddppk1 contain full-length DdPPK1 and its GFP fusion, respectively; pTX-372-Ddppk1 and pTX-gfp-373-Ddppk1 contain truncated DdPPK1 and its GFP fusion, respectively. See Materials and Methods for detail. Data are based on the average values of three independent determinations.
Fig. 4.
Growth of different strains of D. discoideum on a K. aerogenes lawn. D. discoideum cells (20–50) were mixed with K. aerogenes and plated on SM5 agar. Plaque sizes were measured over a 7-day period.
DdPPK1, which shows a significant homology to EcPPK1, is recognized by the antibody raised against EcPPK1. As shown in Fig. 5A, there is only one positive band at ≈118 kDa detected by Western blot against WT extract, whereas no signal was detected in the mutant extract. Immunofluorescent detection of the enzyme in fixed WT cells indicated that DdPPK1 was dispersed probably in membrane vesicles (Fig. 5B), the same as seen for GFP-fusion protein. No immunofluorescent signal was detected in the mutant cells (data not shown).
Fig. 5.
Cellular localization of DdPPK1. (A) Western blot of anti-EcPPK1 against D. discoideum WT and DdPPK1 mutant (AX2 Ddppk1::bsr) soluble extracts. (B) Immunofluorescent detection of DdPPK1 in WT cells using anti-EcPPK1 antibody. (C) DAPI staining image of the same field as B. (D) The merged image of B and C with anti-EcPPK1 signal in green and DAPI in red. Preimmune serum was tested and was found to be negative.
Functions of DdPPK1 N-terminal Domain.
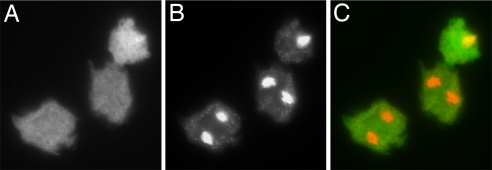
The unique N-terminal extension of 370 aa was essential for DdPPK1 enzymatic activity, cellular localization, and functions. The mutant overexpressing full-length DdPPK1 (containing pTX-DdPPK1) had a very high activity (≈11,000 units/mg in crude lysate), but the N-terminal truncated form (either pTX-372-Ddppk1 or pTX-gfp-373-Ddppk1) showed very little activity in cell extracts (Table 2). The truncated DdPPK1, unlike the full-length DdPPK1, was localized in the cytosol on the basis of the GFP signals of the mutant cells bearing the pTX-gfp-373-Ddppk1 (Fig. 6) rather than in the membrane systems. The truncated DdPPK1 failed to complement the mutant for growth on a K. aerogenes lawn, whereas full-length DdPPK1 complemented the mutant as measured by plaque size (Fig. 4).
Fig. 6.
Cellular location of N-terminal truncated DdPPK1. Log-phase mutant cells containing pTX-gfp-373-Ddppk1 were fixed on cover slips, stained with DAPI, and visualized under microscope. (A) GFP signal of N-terminal-truncated DdPPK1. (B) DAPI staining image of the same field as A. (C) Merged image of A and B with GFP in green and DAPI staining in red.
DdPPK1 Mutant Cells Failed to Complete Cytokinesis.
In addition to its functions in development and phagocytosis (8), DdPPK1 plays a role in the late stages of cytokinesis and cell division. Unlike WT cells, which were almost all mononucleated, ≈30% of DdPPK1 mutant cells were multinucleated when grow on surface in HL5 medium (Table 3). Similar result was obtained with cells in suspension.
Table 3.
The mutant elevated has multinucleated cell populations
| Nuclei per cell* | WT |
DdPPK1 mutant |
||
|---|---|---|---|---|
| Cell no. | % | Cell no. | % | |
| 1 | 619 | 96 | 381 | 68 |
| 2 | 27 | 4 | 172 | 31 |
| 3 | 0 | 0 | 6 | 1 |
*Numbers of nuclei per cell were counted based on DAPI staining of fixed mid-log phase cells that were grown on surface in HL5 medium.
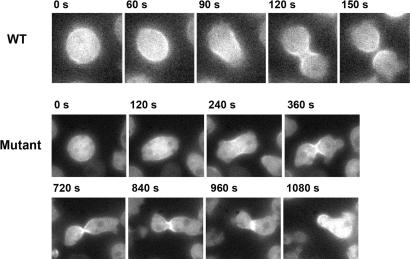
To examine the different stages in cytokinesis, cells were transformed with a myosin II-GFP-expressing plasmid and the process was followed by live-cell microscopy. WT cells completed cell division in <3.5 min after anaphase (Fig. 7), whereas DdPPK1mutant cells took longer, and 16 of 39 cells observed failed to divide. Although the typical myosin II location in the cleavage furrow was seen in all cells, the furrow ingression in the mutant failed to progress to completion (Fig. 7). After 10–25 (average, 17) min, daughter cells of the mutant fused and became mostly binucleated. Also observed were four binucleated parent cells that completed the first round of cytokinesis but failed in the next to become either two binucleated daughter cells or a mononucleated and a trinucleated cell. Of the 19 cells that divided normally, the time interval between anaphase and the end of cytokinesis was between 3 and 10 min, on an average about twice the time taken by the WT cells.
Fig. 7.
Cytokinesis of mutant cells (AX2 Ddppk1::bsr) expressing GFP-myosin II. Time-lapse images show the localization of GFP-myosin II. The WT cell underwent cytokinesis in 150 s. The mutant cell showed normal GFP-myosin II localization and initiation of cleavage furrow ingression. However, it did not complete cleavage furrow ingression and cytokinesis in 18 min (observed in 16 of 39 cells imaged).
Discussion
The principal bacterial enzyme for the synthesis of inorganic poly P is PPK1. This enzyme is widely conserved in bacteria and serves a number of essential roles (1). DdPPK1 is shown to be essential in development, sporulation, and predation (8). Here, we report the purification, characterization, and cellular localization of DdPPK1 and a unique role in cytokinesis.
Although DdPPK1 activity in crude lysate was stimulated by the addition of poly P in the reaction mixture, the effect was very much reduced in partially purified enzyme. This suggests that poly P is required not as a primer, but as a stabilizer, to neutralize the contaminating exopolyphosphatase activity present in crude extract or in partially purified enzyme preparation. So far, no PPK activity from any source has been shown to require or use a primer.
The DdPPK1 sequence reveals an N-terminal domain of 370 aa with no homology to any known protein (SI Fig. 8). The N-terminal truncated DdPPK1 still contains the residues found in bacterial PPK1 for autophosphorylation, ATP binding, and poly P synthesis. However, a mutant lacking the N-terminal sequence is enzymatically inactive and is not localized in the cell as in the WT. Of comparative interest is the PPK1 of P. aeruginosa, which contains an extra N-terminal sequence of 80 aa compared with the enzyme in E. coli (SI Fig. 8), which is not essential for activity (K. Ishige, N.N.R., and A.K., unpublished data).
The molecular size of the isolated DdPPK1 is ≈720 kDa, presumably a hexamer of the 118-kDa subunit (Figs. 1 and 2). In the presence of poly P or ATP, the size increases, probably due to oligomerization. A similar capacity for oligomerization was observed with the hexameric P. aeruginosa PPK1 but not with EcPPK1, which lacks an N-terminal extension (K. Ishige and A.K., unpublished data). These properties of a PPK (oligomerization and cellular localization) are reminiscent of the remarkable DdPPK2 of Dictyostelium (19), which polymerizes into an actin-like filament concurrent with the synthesis of poly P from ATP. Experiments using an antibody against EcPPK1 showed that DdPPK1 was probably membrane-associated (Fig. 5). Even though the pattern of signal distribution reminds one of mitochondrial localization (21), the web-based programs Signal2.0P and Target1.1 (22) do not provide enough evidences for DdPPK1 to be mitochondrial. Moreover, Marchesini et al. found that poly P in D. discoideum is located in acidocalcisomes, the mass-dense granules containing large amounts of phosphorus, magnesium, and calcium (23). It would be interesting to know whether DdPPK1 localizes in acidocalcisomes. This could be achieved by colocalization studies with the vacuolar pyrophosphatase. Therefore, further experiments need to be performed to establish precise cellular location of DdPPK1. Beyond the cellular functions of DdPPK1 reported in ref. 8, we have observed an important role for DdPPK1/poly P in different stages of cytokinesis (Fig. 7 and Table 3). Mutant cells failed either to divide or separate. Those that did took longer time and were likely to be multinucleated. This phenotype of incomplete cleavage furrow-ingression resembled that observed in the clathrin mutant of D. discoideum (24) and in a mutant of kinesin-like protein CHO1 of mammalian cells (25, 26). As with several other cellular events that depend on poly P, the molecular basis for its actions in cytokinesis remain to be determined.
The enzyme DdPPK1 is the first eukaryotic PPK1 studied so far, and it is likely that many will be discovered. One such candidate is in the yeast Candida humicola, in which a PPK1 activity resembling bacterial PPK1 was observed, and a DNA fragment with 34% homology to EcPPK1 in the deduced protein sequence was cloned (15). Furthermore, some photosynthetic eukaryotes (Physcomitrella patens subsp. patens, European Molecular Biology Laboratory accession no. AJ 876529.1) and moss were shown to possess DNA sequences partially homologous to EcPPK1.
Materials and Methods
Reagents.
The following reagents were used: blastocidin (Invitrogen, Carlsbad, CA); G418 (Nalge Nunc, Rochester, NY); ATP (Roche Molecular Biochemicals, Indianapolis, IN); [γ-32P]ATP (Amersham Biosciences, Piscataway, NJ); protein size markers (Bio-Rad, Hercules, CA); polyethylimine-cellulose-F TLC plates (Merck, Darmstadt, Germany); restriction enzymes (New England Biolabs, Ipswich, MA); poly P (types 15 and 75), apyrase, and common chemicals (Sigma–Aldrich, St. Louis, MO); poly P35, poly P50, poly P300, and poly P750 (A. Kornberg laboratory).
Cells and Growth Conditions.
D. discoideum strains include WT (AX2) and Ddppk1 mutant AX2M1 [AX2 Ddppk1::bsr (blasticidin resistance)] (8). All strains were grown at 21°C in HL5 medium (27). Cells were also grown in association with K. aerogenes on SM5 agar plates (28). Antibiotics (blasticidin, 5 μg/ml; G418, 10 μg/ml) were added to the media wherever necessary.
Assay of DdPPK1 Activity.
The assay was performed as described in ref. 8 with minor modifications. The assay mixture contained 50 mM Hepes-KOH (pH 7.2), 80 mM (NH4)2SO4, 1 mM poly P75 (in phosphate residues), 5 mM MgCl2, and 2 mM [γ-32P]ATP. One unit of enzyme is defined as the amount incorporating 1 pmol of phosphate into poly P per min at 37°C.
Isolation of DdPPK1.
D. discoideum WT cells transformed with pTX-Ddppk1 were grown to late log phase, harvested, and the cells were suspended in 100 ml of TED buffer (50 mM Hepes, pH 7.4/1 mM EDTA/DTT) with 200 mM KCl. The cells were lysed by freeze thawing and subjected to ultracentrifugation at 88,000 × g for 1 h at 4°C. The supernatant was filtered through a 0.22-μm filter, loaded onto a Heparin column, and eluted with TED buffer containing an increased gradient of KCl. The fractions with peak DdPPK1 activity were collected and diluted with TED buffer to make the KCl concentration of ≈200 mM. The diluted sample was passed through a DEAE column. The flow-through (FT) fractions were then separated with a Q column.
Size-Exclusion Chromatography.
The native molecular mass of DdPPK1 was determined on a TSK G3000SWXL column (TosoHaas, Tokyo, Japan) in buffer A [50 mM Hepes-KOH, pH 7.4/0.5 mM EDTA/1 mM DTT/0.01% CHAPS/150 mM Na2SO4], buffer B (buffer A plus 10 mM poly P75), or buffer C (buffer A plus 2 mM ATP) at a flow rate of 0.5 ml/min in a PerkinElmer (Waltham, MA) 888 Pechrom HPLC system at 4°C. Marker proteins were detected by UV absorption (wave length 280 nm). Fractions were assayed for DdPPK1 activity.
Urea-PAGE Analysis of the Poly P Product.
The enzyme assay was performed with purified DdPPK1. Products were precipitated from the reaction mixture with 70% (vol/vol) 2-propanol and treated with apyrase (1units/ml at 37°C for 30 min) to remove ATP. An aliquot of the purified product was treated with yeast exopolyphosphatase (29). Electrophoresis was performed on a 20% urea–polyacrylamide gel containing 7 M urea at 600 V for 3 h; the gel was exposed to a PhosphorImager screen (Molecular Dynamics, Sunnyvale, CA).
Truncation and GFP-Fusion Construction.
On the basis of the same strategy used for overexpression of full-length DdPPK1 (8), pTX-372-Ddppk1 was constructed for expressing the truncated form in which the N-terminal region of 372 aa was deleted. Briefly, a 0.35-kb-actin-15-promoter was amplified from pTX-gfp (30) by using primers P1 (8) and P15-DK372 (5′-CATTCTTGAACCACTCATTTTTTAAGCTTG-3′); the truncated fragment of DdPPK1 was amplified from WT genomic DNA with primers, DK1-P15–372 (5′-CAAGCTTAAAAAATGAGTGGTTCAAGAATG-3′) and DK1-Xba-L4 (8). The two PCR products were purified and mixed as templates for the second-round PCR with primers P1 and DK1-Xba-L4. The product was digested with SalI and XbaI and inserted into pTX-gfp to obtain pTX-372-Ddppk1, which was then transferred into the Ddppk1 mutant (AX2M1) and selected as described in ref. 8.
The GFP fusion of full length and truncated DdPPK1 were constructed downstream of gfp in pTX-gfp vector (30). The full-length DdPPK1 fragment was amplified with DK1-Sal-U1 (5′-GGTCGACATTGCATTGTATTTTCAGACTA-3′) and DK1-Xba-L4 (8), was digested with SalI and XbaI, and was then inserted into pTX-gfp to obtain pTX-gfp-Ddppk1. The truncated DdPPK1 fragment was amplified with primers DK1-Xho-373 (5′-ACTCGAGGTTCAAGAATGTTTTTTAATAGG-3′) and DK1-Xba-L4 (8), in which the first 373 amino acids from N-terminal of DdPPK1 were deleted. The fragment was digested with XhoI and XbaI, then inserted into pTX-gfp to obtain pTX-gfp-373-Ddppk1. These plasmids were then transformed into both WT and the Ddppk1 mutant.
Transformation of D. discoideum.
Ddppk1 mutant (AX2M1) cells (8) were grown in HL5 medium in 9-cm dishes to mid-log phase, pelleted by centrifugation at 2,000 × g for 5 min at 4°C, washed with electroporation buffer (10 mM sodium phosphate, pH 7.0; 50 mM sucrose), and resuspended in a proper volume of the same buffer. Electroporation was performed in a 4-mm cuvette <1.3 KV and 3.0 μF. Cells were cultured in 10 ml of HL5 medium for 20 h, and selective antibiotics were added to the medium for selection.
Nuclear Number Studies.
WT and mutant cells were grown on coverslips in HL5 medium to mid-log phase. The cells were fixed in methanol at −15°C for 5 min and then washed three times with PBS containing 0.05% Tween 20. After staining with DAPI (10 μg/ml), nuclei were counted on the basis of DAPI signal by using a Nikon fluorescent microscope (Tokyo, Japan).
Western Blot Analysis.
After SDS/PAGE, the proteins were transferred onto nitrocellulose membranes by using a Transblot apparatus (Bio-Rad) blocked with 5% (wt/vol) nonfat milk in PBS and incubated overnight at 4°C. A polyclonal rabbit anti-EcPPK1 antibody was diluted (×10,000) in the blocking buffer and incubated with blots at room temperature for 60 min. The nitrocellulose membranes were washed three times for 20 min each with PBS containing 0.05% Tween 20 before the addition of a 1:10,000 dilution of goat anti-rabbit IgG peroxidase antibody conjugate in blocking buffer for 30 min.
Detection of PPK1 by Immunofluorescence or GFP Signal.
WT and mutant cells grown on cover slips were fixed for 5 min at −15°C in methanol and washed three times with PBS containing 0.05% Tween 20. The coverslips were then blocked in PBS containing 0.1% BSA for 30 min at 37°C. After incubation for 30 min with anti-PPK1 antibody (1:200), the coverslips were thoroughly rinsed with PBS containing 0.05% Tween 20. Further incubation was performed with FITC-conjugated secondary antibody (1:1,000). The coverslips were placed on a glass slide and examined for immunofluorescence by using a Nikon fluorescent microscope equipped with a 100× objective. Fluorescence images were obtained by using a green filter for GFP and a blue filter for DAPI. Pictures were captured with a CDD digital camera and processed with MetaMorph software (Universal Imaging Corporation, Downingtown, PA).
Cells containing GFP fusion proteins were grown on cover slips and fixed in methanol. After staining with DAPI, the signals were observed the same way with immunofluorescence.
Fluorescent Live-Cell Microscopy.
For live-cell imaging, the cells carrying GFP-myosin II (transformed with plasmid p100, gift from J. A. Spudich, Stanford University, Stanford, CA) were cultivated at 22°C in optical coverglass chambers (Lab-Tek, Naperville, IL), and the HL5 medium was replaced by imaging buffer [20 mM Mes-NaOH (pH 6.8)/0.2 mM CaCl2/2 mM Mg2SO4] (31). Cells were examined with a 64× 1.3 numerical aperture objective on an Axiovert microscope (Zeiss, Jena, Germany). Images were collected at 30-s intervals with MetaMorph software and processed with Photoshop (Adobe Systems).
Supplementary Material
Acknowledgments
We thank Dr. James A. Spudich for valuable advice on experimental design, and Drs. Hans M. Warrick and Gandikota S. Lakshmikanth for help in fluorescent live-cell microscopy and discussions. This work was supported by grants from the National Institutes of Health.
Abbreviations
- poly P
inorganic polyphosphate
- PPK
polyphosphate kinase
- DdPPK1
Dictyostelium discoideum PPK1
- EcPPK1
Escherichia coli PPK1
- Pi
orthophosphate.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF176830).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706847104/DC1.
References
- 1.Kornberg A, Rao NN, Ault-Riché D. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 2.Rao NN, Kornberg A. J Bacteriol. 1996;178:1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 4.Rashid MH, Rao NN, Kornberg A. J Bacteriol. 2000;182:225–227. doi: 10.1128/jb.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rashid MH, Kornberg A. Proc Natl Acad Sci USA. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X, Rao NN, Kornberg A. Proc Natl Acad Sci USA. 2004;101:17061–17065. doi: 10.1073/pnas.0407787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rashid MN, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. Proc Natl Acad Sci USA. 2000;97:9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Gómez-García MR, Brown MRW, Kornberg A. Proc Natl Acad Sci USA. 2005;102:2731–2735. doi: 10.1073/pnas.0500023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Rao NN, Shiba T, Kornberg A. Proc Natl Acad Sci USA. 2005;102:13416–13420. doi: 10.1073/pnas.0506520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith SA, Mutch NJ, Baskar D, Rohloff P, Docampo R, Morrissey JH. Proc Natl Acad Sci USA. 2006;103:903–908. doi: 10.1073/pnas.0507195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Fraley CD, Faridi J, Kornberg A, Roth RA. Proc Natl Acad Sci USA. 2003;100:11249–11254. doi: 10.1073/pnas.1534805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reusch RN, Sadoff HL. Proc Natl Acad Sci USA. 1988;85:4176–4180. doi: 10.1073/pnas.85.12.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pavlov E, Zakharian E, Bladen C, Diao CT, Grimbly C, Reusch RN, French RJ. Biophys J. 2005;88:2614–2625. doi: 10.1529/biophysj.104.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahn K, Kornberg A. J Biol Chem. 1990;265:11734–11739. [PubMed] [Google Scholar]
- 15.McGrath JW, Kulakova AN, Kulakova LA, Quon JP. Res Microbiol. 2005;156:485–491. doi: 10.1016/j.resmic.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Ishige K, Kornberg A. Proc Natl Acad Sci USA. 2002;99:16684–16688. doi: 10.1073/pnas.262655299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Huang W, Lee SSK, Xu W. EMBO Rep. 2005;6:681–687. doi: 10.1038/sj.embor.7400448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumble KD, Ahn K, Kornberg A. Proc Natl Acad Sci USA. 1996;93:14391–14395. doi: 10.1073/pnas.93.25.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gómez-García MR, Kornberg A. Proc Natl Acad Sci USA. 2004;101:15876–15880. doi: 10.1073/pnas.0406923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz de la EM, Pollard TD. Biochemistry. 1994;33:14387–14392. doi: 10.1021/bi00252a003. [DOI] [PubMed] [Google Scholar]
- 21.van Esa S, Wesselsb D, Sollb DR, Borleisa J, Devreotes PN. J Cell Biol. 2001;152:621–632. doi: 10.1083/jcb.152.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emanuelsson O, von Heijne G. Biochim Biophys Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- 23.Marchesini N, Ruiz FA, Vieira M, Docampo R. J Biol Chem. 2002;277:8146–8153. doi: 10.1074/jbc.M111130200. [DOI] [PubMed] [Google Scholar]
- 24.Gerald NJ, Damer CK, O'Halloran TJ, Lozanne AD. Cell Motil Cytoskel. 2001;48:213–223. doi: 10.1002/1097-0169(200103)48:3<213::AID-CM1010>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 25.Kuriyama R, Gustus C, Terada Y, Uetske Y, Matuliene J. J Cell Biol. 2002;156:783–790. doi: 10.1083/jcb.200109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matuliene J, Kuriyama R. Mol Biol Cell. 2002;13:1832–1845. doi: 10.1091/mbc.01-10-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spudich JA. Methods Cell Biol. 1982;25:359–364. doi: 10.1016/s0091-679x(08)61433-8. [DOI] [PubMed] [Google Scholar]
- 28.Loomis WF. Dictyostelium discoideum: A Developmental System. New York: Academic; 1975. [Google Scholar]
- 29.Wurst H, Shiba T, Kornberg A. J Bacteriol. 1995;177:898–906. doi: 10.1128/jb.177.4.898-906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levi S, Polyakov M, Egelhoff TT. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- 31.Lakshmikanth GS, Warrick HM, Spudich JS. Proc Natl Acad Sci USA. 2004;101:16519–16524. doi: 10.1073/pnas.0407304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.