Summary
Mammalian orthoreoviruses (reoviruses) are highly tractable experimental models for studies of double-stranded (ds) RNA virus replication and pathogenesis. Reoviruses infect respiratory and intestinal epithelium and disseminate systemically in newborn animals. Until now, a strategy to rescue infectious virus from cloned cDNA has not been available for any member of the Reoviridae family of dsRNA viruses. We report the generation of viable reovirus following plasmid transfection of murine L929 (L) cells using a strategy free of helper virus and independent of selection. We used the reovirus reverse genetics system to introduce mutations into viral capsid proteins σ1 and σ3 and to rescue a virus that expresses a green fluorescent protein (GFP) transgene, thus demonstrating the tractability of this technology. The plasmid-based reverse genetics approach described here can be exploited for studies of reovirus replication and pathogenesis and used to develop reovirus as a vaccine vector.
Keywords: MICROBIO
Introduction
Reoviruses are members of the Reoviridae family, which includes several genera that cause disease in humans and animals. Chief among these are the rotaviruses, which are the most common cause of viral gastroenteritis in human infants (Kapikian et al., 2001). Reoviruses infect the respiratory and gastrointestinal tracts of virtually all mammals, including humans (Tyler, 2001). However, disease associated with reovirus infection occurs primarily in the very young (Tyler et al., 2004). Reoviruses are highly virulent in newborn mice, the preferred experimental system for studies of reovirus pathogenesis, and produce injury to a variety of host tissues, including the central nervous system (CNS), heart, and liver (Tyler, 2001).
Reoviruses contain a genome of 10 dsRNA gene segments enclosed in two concentric protein shells, termed outer capsid and core. Reovirus infection is initiated by viral attachment to host cells via the filamentous attachment protein σ1 (Furlong et al., 1988). The σ1 protein engages cell-surface carbohydrate (Chappell et al., 1997, Chappell et al., 2000) and junctional adhesion molecule-A (JAM-A) (Barton et al., 2001b, Campbell et al., 2005), an integral component of intercellular tight junctions (Martin-Padura et al., 1998). Following attachment to the cell surface, reovirus internalization is mediated by β1 integrins (Maginnis et al., 2006), most likely via clathrin-dependent endocytosis (Ehrlich et al., 2004). In the endocytic compartment, viral outer-capsid proteins σ3 and μ1/μ1C are cleaved by acid-dependent cysteine proteases (Baer and Dermody, 1997, Ebert et al., 2002), resulting in generation of infectious subvirion particles (ISVPs) (Borsa et al., 1981). During ISVP formation, σ3 is removed and a hydrophobic conformer of μ1/μ1C is exposed, facilitating endosomal membrane penetration and delivery of transcriptionally active reovirus core particles into the cytoplasm (Chandran et al., 2002, Odegard et al., 2004), where the remainder of the replication cycle occurs.
With the exception of dsRNA viruses, a plasmid-based reverse genetics system exists for all major groups of animal RNA viruses, including bornaviruses, bunyaviruses, coronaviruses, flaviviruses, orthomyxoviruses, paramyxoviruses, picornaviruses, and rhabdoviruses (Table S1 in the Supplemental Data available with this article online). Despite extensive efforts in several laboratories, generation of an animal dsRNA virus entirely from cloned cDNAs has not been achieved. This critical technological gap is perhaps the single most important limitation to studies of these viruses. Previous efforts on reovirus and rotavirus reverse genetics have resulted in entirely RNA-based (Roner et al., 1997) or partially plasmid-based (Komoto et al., 2006) systems that permit replacement of one or two viral genes. These approaches have been used to rescue temperature-sensitive reovirus strains (Roner et al., 1997), define packaging signals in reovirus RNAs (Roner and Steele, 2007), and isolate rotaviruses containing engineered changes in the viral attachment protein (Komoto et al., 2006). However, neither the reovirus nor rotavirus reverse genetics systems in their current configurations permit selective introduction and recovery of desired mutations in each viral gene segment.
We report the development of an entirely plasmid-based reverse genetics system for mammalian reovirus in which viable viruses are generated from cloned cDNAs. Neither helper virus nor coexpression of viral replication proteins is required for recovery of wild-type (WT) virus or engineered viral mutants. Point mutations introduced into viral capsid proteins σ1 and σ3 were used to define sequences that govern susceptibility to cleavage by intestinal proteases. We also recovered a recombinant virus that expresses green fluorescent protein (GFP) by replacement of the σ3 open reading frame (ORF) with GFP. The establishment of plasmid-based reverse genetics for reovirus will allow heretofore technically unapproachable problems in dsRNA virus biology to be studied, provide a platform for development of analogous marker rescue systems for other segmented dsRNA viruses, and foster exploration of reovirus as a vaccine vector to elicit protective immunity against a variety of mucosal pathogens.
Results
Generation of Viable Reovirus from Cloned cDNA
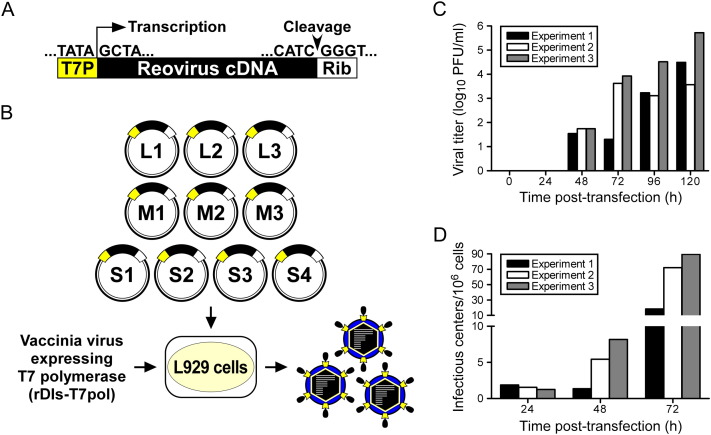
To generate recombinant reovirus from cloned cDNAs, plasmids encoding each of the ten viral gene segments were engineered to facilitate transcription of full-length viral mRNAs under control of the bacteriophage T7 RNA polymerase promoter, which directs transcription initiation preferentially from a juxtaposed guanosine residue (Milligan et al., 1987). As all reovirus (+)-sense RNAs are terminated with a 5′ guanosine (Furuichi et al., 1975a, Furuichi et al., 1975b), plasmid-generated transcripts are anticipated to possess native 5′ ends (Roner and Joklik, 2001) ( Figure 1A). Murine L929 fibroblast (L) cells, which efficiently support reovirus replication (Barton et al., 2001a), were infected with the attenuated, T7 RNA polymerase-expressing vaccinia virus strain rDIs-T7pol 1 hr prior to transfection with the ten reovirus cDNA plasmids (Figure 1B). Nascent transcripts were synthesized with the hepatitis delta virus (HDV) ribozyme fused to the 3′ terminus, which is expected to generate a native 3′ end upon autocatalytic removal (Roner and Joklik, 2001) (Figure 1A). Thus, this expression strategy should yield ten unique reovirus mRNA species competent to complete all steps in the viral RNA life cycle. Accordingly, rescued viruses were recovered from cell-culture supernatants by plaque assay on L cell monolayers (Figure 1C).
Figure 1.
Experimental Strategy to Generate Reovirus from Cloned cDNA
(A) Prototype reovirus gene segment cDNA in plasmid. Cloned cDNAs representing each of the ten full-length reovirus RNA gene segments are flanked by the bacteriophage T7 RNA polymerase promoter (T7P) and the antigenomic hepatitis delta virus (HDV) ribozyme (Rib).
(B) Schematic of approach. The ten reovirus cDNA constructs are transfected into murine L cells expressing T7 RNA polymerase from recombinant vaccinia virus strain rDIs-T7pol, which is replication defective. Nascent transcripts correspond to viral mRNAs containing the native 5′ end. Self cleavage by the HDV ribozyme generates the native viral 3′ end. Following 5 days of incubation, transfected cells are lysed by freeze-thaw, and viable viruses rescued from cloned cDNAs are isolated by plaque assay using L cells.
(C) Kinetics of virus production following plasmid transfection of L cells. Cells were transfected with plasmid DNA according to the protocol in (B) and lysed at the intervals shown. Viral titers in cell lysates were determined by plaque assay.
(D) Infectious center assay following plasmid transfection of L cells. Cells were transfected with plasmid DNA, trypsinized at the intervals shown posttransfection, washed, counted, diluted, and applied directly to monolayers of untreated L cells. The number of the infectious centers was determined by plaque assay.
To ensure that viruses isolated following plasmid transfection represented single clones, and to preclude contamination of reovirus stocks by rDIs-T7pol, all viruses were isolated by plaque purification using L cell monolayers. rDIs-T7pol is replication defective and produces no detectable growth in mammalian cells (Ishii et al., 2002). Concordantly, no viral plaques arose on L cell monolayers when the cotransfections were prepared with nine of the ten reovirus marker-rescue plasmids (data not shown). Furthermore, vaccinia virus proteins were not detected by immunoblot analysis of second-passage stocks of plaque-purified reoviruses (data not shown).
Infectious center assays were performed to assess the efficiency of reovirus infection in plasmid-transfected L cells. At 24–48 hr posttransfection, times corresponding to the primary round of infection, eight or fewer infectious centers per 106 cells were detected (Figure 1D). The number of infectious centers increased substantially between 48 and 72 hr posttransfection to 18 to 90 per 106 cells, suggesting that a secondary round of infection had ensued by the 72 hr time point. Viral titer in transfection lysates was below the limit of detection (∼10 PFU/ml) at 24 hr but was detectable at 48 hr and increased logarithmically at time points thereafter (Figure 1C). The ratio of viral PFU in transfection lysates to infectious centers was ∼10 to 100 at 48 and 72 hr. These results indicate that initiation of productive reovirus replication from transfected plasmids is inefficient, with approximately 1 cell per 105 to 106 cells giving rise to, on average, 10 to 100 viable virus particles that establish infection of the culture. In our experiments, yields of WT virus 5 days after plasmid transfection are regularly in the range of 104–106 PFU/ml (Figure 1C).
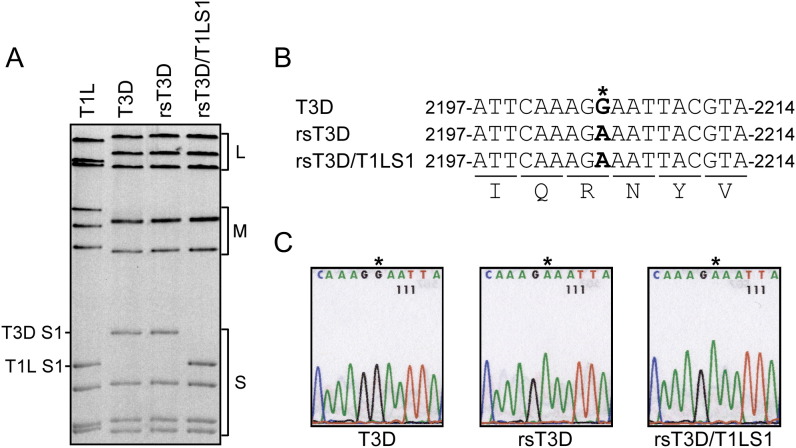
Separation of reovirus genomic dsRNA using SDS-PAGE produces unique electrophoretic patterns that can be used to discriminate different viral strains (Barton et al., 2001a). To confirm that viruses isolated using the plasmid-based rescue procedure contained the expected combination of gene segments, genomic dsRNA isolated from recombinant strain (rs) T3D and rsT3D/T1LS1 was resolved in SDS-polyacrylamide gels and visualized by ethidium bromide staining ( Figure 2A). The electropherotype of rsT3D was indistinguishable from that of strain T3D, the origin of the cloned cDNA sequences used to generate rsT3D. Likewise, rsT3D/T1LS1 displayed an electropherotype consistent with nine cloned gene segments derived from T3D and a single cloned gene segment, S1, derived from strain T1L. To exclude the possibility of contamination, a silent point mutation, G to A at nucleotide 2205, was introduced into the L1 gene of all virus strains generated from cloned cDNAs (Figure 2B). This change has not been observed in any reported T3D L1 sequence (Wiener and Joklik, 1989) and serves as a signature for viruses derived through plasmid-based rescue. As anticipated, sequence analysis of rsT3D and rsT3D/T1LS1 revealed the expected G to A substitution (Figure 2C), confirming the plasmid origins of these isolates.
Figure 2.
Rescue of rsT3D and rsT3D/T1LS1
(A) Electropherotypes of T1L, T3D, rsT3D, and rsT3D/T1LS1. Viral dsRNA was extracted from purified virions and electrophoresed in an SDS-polyacrylamide gel, followed by ethidium bromide staining to visualize viral gene segments. Size classes of gene segments (L, M, S) are indicated.
(B) Recombinant viruses contain a novel mutation in the L1 gene. The single nucleotide difference in L1 unique to rsT3D and rsT3D/T1LS1 is shown in the alignment as an asterisk. The G→A substitution at position 2205 is a signature change engineered into the cloned T3D L1 cDNA used for marker rescue.
(C) Sequence analysis of L1 gene RT-PCR products from rescued reoviruses. A fragment of the L1 gene was amplified by one-step RT-PCR performed using viral dsRNA extracted from purified virions of T3D, rsT3D, and rsT3D/T1LS1. Products were subjected to direct sequence analysis and compared to the L1 sequence of T3D. Shown are sequence chromatograms demonstrating G→A substitution at position 2205 of the rsT3D and rsT3D/T1LS1 L1 genes.
Characterization of Reoviruses Generated by Plasmid Transfection
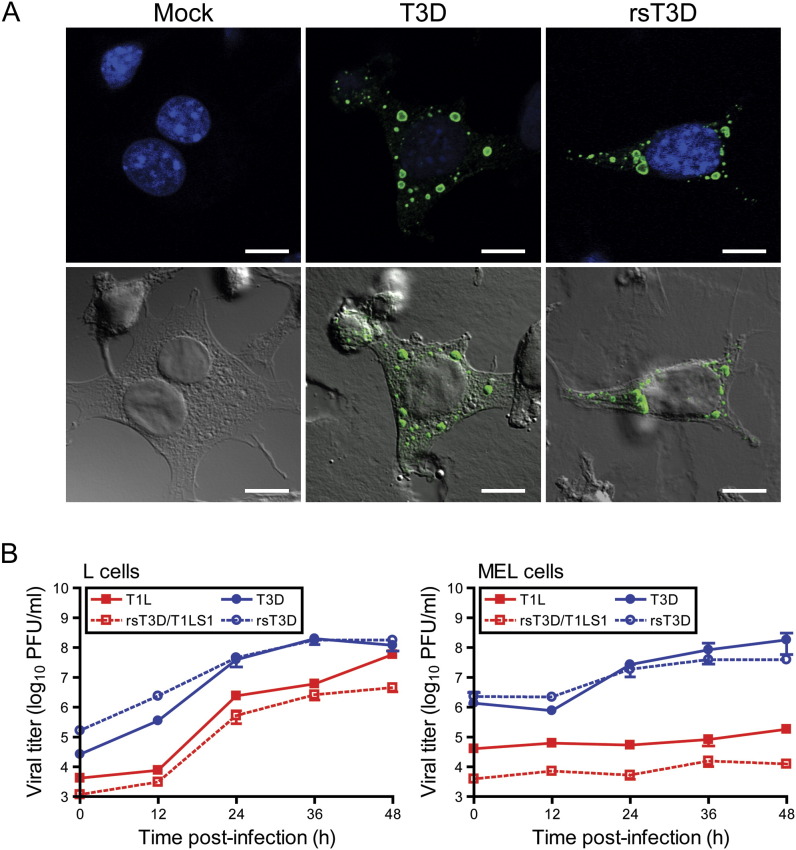
Reoviruses replicate and assemble within cytoplasmic inclusions in infected cells (Fields, 1971). Viral inclusions contain dsRNA (Silverstein and Schur, 1970), viral proteins (Fields, 1971), and both complete and incomplete viral particles (Fields, 1971). Reovirus strain T3D forms large globular inclusions that localize to the perinuclear space (Parker et al., 2002). To determine whether rsT3D forms viral inclusions in a manner similar to native T3D, cells were infected with T3D and rsT3D and processed 18 hr postinfection for image analysis by confocal microscopy ( Figure 3A). Both T3D and rsT3D formed morphologically indistinguishable large globular inclusions that were localized to the perinuclear compartment. We conclude that recombinant rsT3D recapitulates a hallmark of native T3D infection in cultured cells.
Figure 3.
Infectivity of Native and Recombinant Reoviruses
(A) Immunofluorescence of cells infected with T3D and rsT3D. L cells were mock infected or infected with either T3D or rsT3D, stained at 18 hr postinfection with anti-μNS antiserum to visualize reovirus inclusions (green) and TO-PRO3 to visualize nuclei (blue), and imaged by confocal laser scanning microscopy. Representative digital fluorescence (top panel) and DIC images (bottom panel) for mock-, T3D-, and rsT3D-infected cells are shown. Scale bar, 10 μM.
(B) One-step growth curve for T1L, T3D, rsT3D, and rsT3D/T1LS1 in L cells (left) and MEL cells (right). Cells were infected with virus, incubated for the intervals shown, and lysed by freeze-thaw. Viral titers in cell lysates were determined by plaque assay. The results are presented as the mean viral titers for triplicate experiments. Error bars indicate SD.
To confirm that the recombinant viruses exhibit replication kinetics similar to the native strains, T1L, T3D, rsT3D, and rsT3D/T1LS1 were tested for infection of L cells and MEL cells (Figure 3B). L cells support replication of all reovirus strains tested in our laboratory. In contrast, MEL cells support replication of only sialic acid-binding reovirus strains (Rubin et al., 1992, Chappell et al., 1997). T1L, rsT3D/T1LS1, T3D, and rsT3D produced ∼1000-fold yields of viral progeny in L cells. However, only sialic acid-binding strains T3D and rsT3D were capable of efficiently infecting MEL cells, producing yields in each case of ∼100-fold, whereas strains T1L and rsT3D/T1LS1 produced minimal yields of viral progeny in these cells (<10-fold). Together, these data indicate that recombinant reoviruses display replication characteristics indistinguishable from native strains.
Susceptibility of Attachment Protein σ1 to Proteolytic Cleavage Attenuates Reovirus Intestinal Infection and Systemic Spread
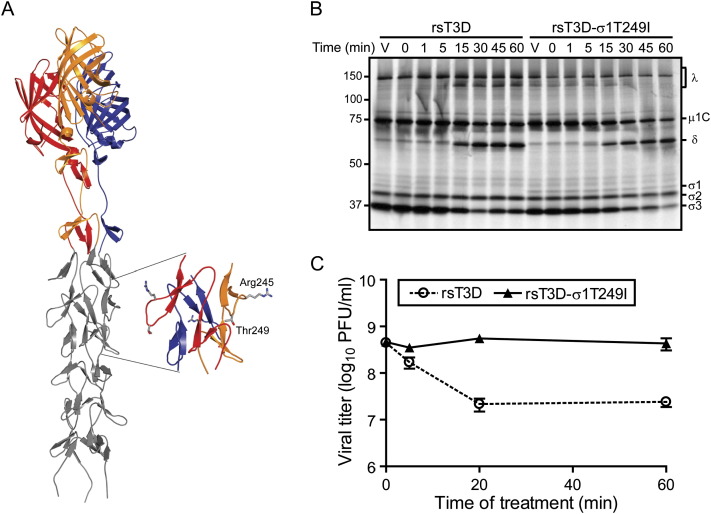
The σ1 protein exhibits strain-specific differences in susceptibility to cleavage following in vitro treatment with intestinal proteases to generate ISVPs (Nibert et al., 1995, Chappell et al., 1998). This difference in cleavage susceptibility segregates with a single amino acid polymorphism in the tail domain of σ1 ( Figure 4A). Strains with a threonine at residue 249 in σ1 are susceptible to cleavage by trypsin after Arg245, whereas those with an isoleucine at residue 249 are resistant to cleavage (Chappell et al., 1998). The importance of sequence polymorphism at residue 249 has been confirmed in studies using expressed protein (Chappell et al., 1998) and recoated core particles (Chandran et al., 2001) but not with intact virions.
Figure 4.
The σ1 Protein of rsT3D-σ1T249I Is Resistant to Trypsin
(A) Model of σ1 generated by adding five β spiral repeats to the N terminus of the crystallized fragment of σ1 (Chappell et al., 2002). The three monomers in the crystallized fragment are shown in red, yellow, and blue; the model is shown in gray. The inset shows an enlarged view of the β spiral region in σ1 that influences susceptibility of the molecule to cleavage by intestinal proteases (Chappell et al., 1998). Side chains of Arg245 and Thr249 are depicted in ball-and-stick representation.
(B) Electrophoretic analysis of viral structural proteins of rsT3D and rsT3D-σ1T249I during treatment with trypsin to generate ISVPs. Purified 35S-methionine-labeled virions were treated with trypsin for the indicated intervals and loaded into wells of a 4%–20% polyacrylamide gradient gel. After electrophoresis, the gel was prepared for fluorography and exposed to film. Samples of untreated virions appear in the lanes labeled “V.” Viral proteins are labeled. Positions of molecular weight standards (in kDa) are indicated. The experiments shown are representative of two performed for each virus.
(C) Infectivity of rsT3D and rsT3D-σ1T249I during treatment with trypsin to generate ISVPs. Purified virions were treated with trypsin at 37°C for the intervals shown. Titers of virus in the treatment mixtures were determined by plaque assay. The results are presented as the mean viral titers for triplicate experiments. Error bars indicate SD.
To determine whether the single Thr-Ile polymorphism at residue 249 in σ1 protein is sufficient to alter σ1 cleavage susceptibility during treatment of virions with intestinal proteases to generate ISVPs, we used plasmid-based rescue to generate rsT3D-σ1T249I, which differs from rsT3D by the presence of an isoleucine in σ1 at residue 249 (Table S2). Purified virions of rsT3D and rsT3D-σ1T249I were treated with trypsin and analyzed by SDS-PAGE. As expected, a digestion pattern consistent with formation of ISVPs (loss of σ3 protein and generation of the δ fragment of μ1C protein) was observed for both viruses (Figure 4B). However, the stability of rsT3D and rsT3D-σ1T249I σ1 proteins differed. The band corresponding to rsT3D σ1 diminished in intensity immediately after trypsin addition until it was eventually undetectable (Figure 4B). However, the rsT3D-σ1T249I σ1 band was intact even after 60 min of digestion. Thus, the T249I polymorphism is an independent determinant of σ1 cleavage susceptibility.
Proteolytic cleavage of σ1 at a site adjacent to Thr249 releases the JAM-A-binding σ1 head domain, leading to diminished viral infectivity (Nibert et al., 1995). To test whether rsT3D and rsT3D-σ1T249I differ in infectivity after protease treatment to generate ISVPs, purified virions of each strain were exposed to trypsin for various intervals, and titers of infectious virus in the treatment mixtures were determined by plaque assay (Figure 4C). As observed with WT T3D in previous experiments (Nibert et al., 1995), rsT3D lost infectious titer rapidly after protease treatment. In contrast, titers of rsT3D-σ1T249I remained relatively stable throughout the assay time course. Loss of infectivity of rsT3D correlated with kinetics of σ1 cleavage (compare Figures 4B and 4C), indicating that changes in viral infectivity after trypsin treatment are governed by the cleavage state of σ1. Furthermore, both phenotypes are linked to a single σ1 polymorphism at amino acid 249.
Reovirus strains T1L and T3D differ in the capacity to infect the murine intestine after peroral (PO) inoculation (Bodkin et al., 1989), a property that segregates with the viral S1 (encoding σ1 and σ1s) and L2 (encoding λ2) genes (Bodkin and Fields, 1989). Exposure of T3D to an intestinal wash results in σ1 cleavage (Chappell et al., 1998), raising the possibility that failure of T3D to infect the intestine is in part attributable to σ1 cleavage susceptibility. To test whether susceptibility of σ1 to proteolytic cleavage is associated with diminished T3D growth in animals, we assessed the capacity of rsT3D and rsT3D-σ1T249I to infect the intestine and disseminate systemically following PO inoculation ( Figure 5A). Newborn mice were inoculated perorally with each strain, and viral titers in the intestine and brain were determined at 4, 8, and 12 days after inoculation. At all time points tested, titers of rsT3D-σ1T249I in the intestine were greater than those produced by rsT3D. Furthermore, rsT3D-σ1T249I produced greater titers in the brain at days 8 and 12 than did rsT3D. However, when inoculated by the intracranial (IC) route, rsT3D and rsT3D-σ1T249I produced equivalent titers (Figure 5B), although rsT3D reached peak titers at earlier time points than did rsT3D-σ1T249I. These findings indicate that a Thr-Ile polymorphism at amino acid 249 in T3D σ1 controls viral growth in the murine intestine and systemic spread to the CNS.
Figure 5.
rsT3D-σ1T249I Infects the Murine Intestine and Disseminates to the CNS
Titers of rsT3D and rsT3D-σ1T249I after either PO (A) or IC (B) inoculation. Mice were inoculated with virus and euthanized at the indicated times postinoculation. Viral titers in organ homogenates were determined by plaque assay. The limit of detection was 100 PFU/ml of organ homogenate. Each data point represents the average viral titer from 3 to 12 mice. Error bars indicate SD.
Regulation of Reovirus Disassembly by a Single Polymorphism in Outer-Capsid Protein σ3
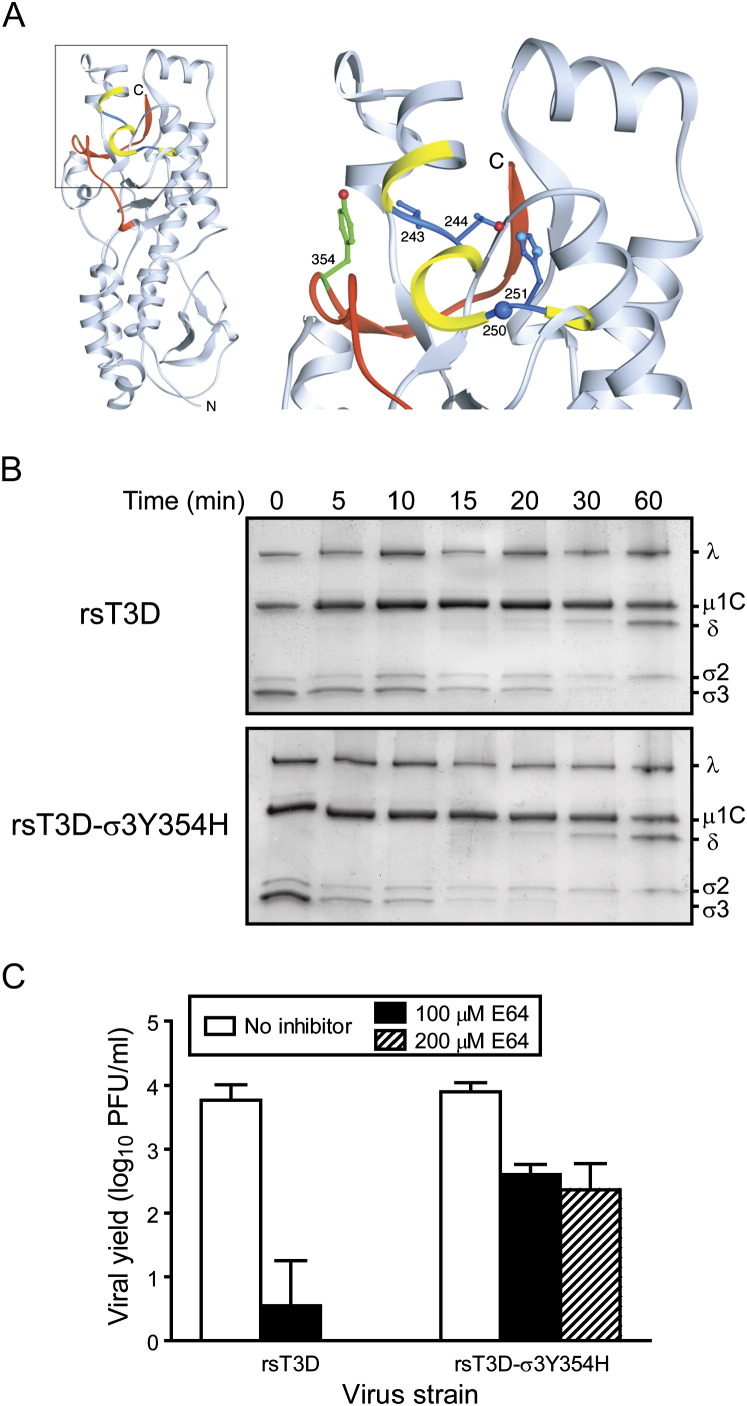
Previous studies identified a tyrosine-to-histidine substitution at amino acid 354 in T3D σ3 as a key regulator of the kinetics of virion-to-ISVP conversion in vitro (Wetzel et al., 1997) and viral resistance to growth inhibition by the cysteine protease inhibitor E64 (Baer and Dermody, 1997, Ebert et al., 2001). Tyr354 is located in the virion-distal lobe of σ3 adjacent to sites in the protein that are cleaved during formation of ISVPs (Ebert et al., 2002) ( Figure 6A). The importance of this residue in viral replication has been deduced by analysis of reassortant viruses containing WT and mutant σ3 proteins (Wetzel et al., 1997, Ebert et al., 2001, Clark et al., 2006) and analysis of ISVPs recoated with WT and mutant forms of σ3 (Wilson et al., 2002, Clark et al., 2006).
Figure 6.
A Single Mutation in Outer-Capsid Protein σ3 Accelerates Proteolytic Disassembly of Reovirus
(A) Crystal structure of T3D σ3 (Olland et al., 2001), in which cathepsin L cleavage sites are depicted in blue between amino acids 243 and 244 and between 250 and 251 (Ebert et al., 2002). Surrounding residues, from amino acids 241 to 253, are shown in yellow. The C-terminal residues of σ3, from amino acids 340–365, are colored red. Tyr354, which is altered in several PI (Wetzel et al., 1997), D-EA (Ebert et al., 2001), and ACA-D viruses (Clark et al., 2006), is shown in green. The virion-distal end of σ3 is at the top of the figure, and the virion-proximal end and N terminus are at the bottom. The inset shows an enlarged view of the boxed region of σ3 using the same color scheme. Side chains of amino acids 243, 244, 250, 251, and 354 are depicted in ball-and-stick representation.
(B) Chymotrypsin treatment of rsT3D and rsT3D-σ3Y354H. Purified virions were treated with chymotrypsin for the indicated intervals and loaded into wells of 10% polyacrylamide gels. After electrophoresis, the gels were stained with Coomassie blue. Viral proteins are labeled. The experiments shown are representative of two performed for each virus.
(C) Growth of rsT3D and rsT3D-σ3Y354H in L cells treated with E64. L cells were preincubated in medium with or without E64 at the concentrations shown. The medium was removed, cells were adsorbed with virus for 1 hr, and fresh medium with or without E64 was added. After 24 hr incubation, viral titers in cell lysates were determined by plaque assay. The results are presented as the mean viral yields, calculated by dividing titer at 24 hr by titer at 0 hr for each concentration of E64, for triplicate experiments. Error bars indicate SD.
To determine whether the Y354H mutation in σ3 is sufficient to confer enhanced virion-to-ISVP conversion and resistance to E64, we generated rsT3D-σ3Y354H (Table S2) and compared this virus to rsT3D for kinetics of σ3 proteolysis following protease treatment in vitro. Virions of each strain were treated with chymotrypsin for various intervals and processed for analysis of viral structural proteins by SDS-PAGE (Figure 6B). Treatment of rsT3D and rsT3D-σ3Y354H virions with chymotrypsin resulted in degradation of σ3 and cleavage of μ1C to form δ, indicative of ISVP formation. Proteolysis of rsT3D-σ3Y354H σ3 during chymotrypsin treatment occurred with substantially faster kinetics than that of rsT3D σ3. This result indicates that amino acid 354 in σ3 protein is an independent determinant of virion susceptibility to proteolytic digestion and likely functions as an autonomous regulator of viral disassembly in cellular endosomes.
The role of σ3 mutation Y354H in virion disassembly in cyto was investigated by quantifying yields of rsT3D and rsT3D-σ3Y354H after 24 hr of growth in L cells treated with 0–200 μM E64 (Figure 6C). Both strains produced yields of ∼1000 fold following growth in untreated cells. However, yields of rsT3D-σ3Y354H were ∼100-fold greater than those of rsT3D following growth in cells treated with 200 μM E64 (the highest concentration tested). Therefore, a single mutation in σ3, Y354H, regulates resistance of reovirus to an inhibitor of cysteine proteases within cellular endosomes.
Transduction of GFP by a Recombinant Reovirus
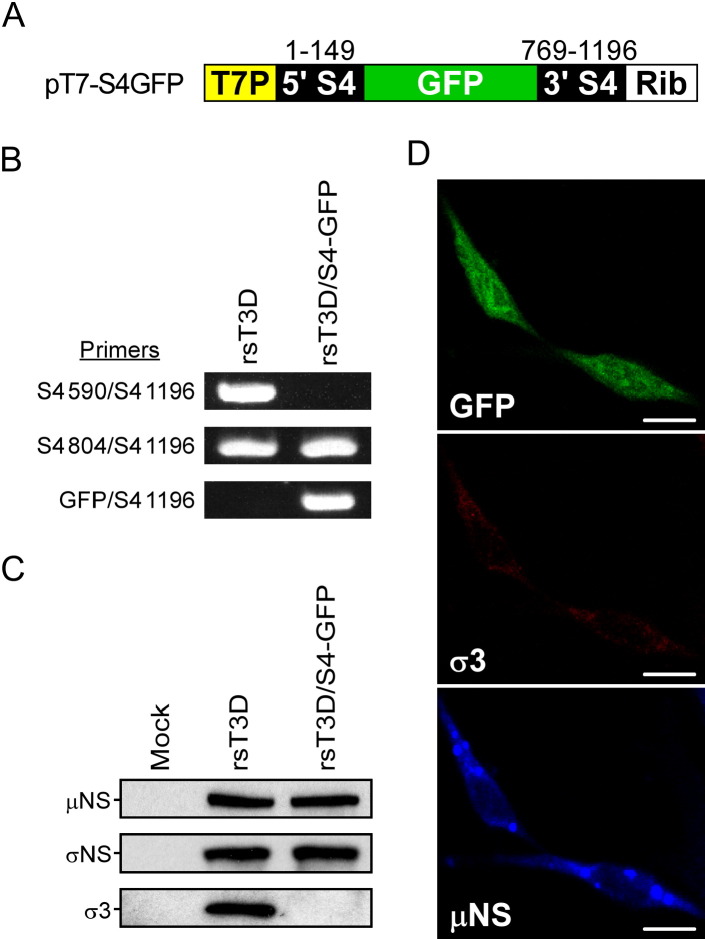
To determine whether reoviruses capable of expressing a foreign gene can be recovered following plasmid transfection, we introduced sequences encoding GFP into the σ3 ORF of the T3D S4 plasmid ( Figure 7A). In this configuration, GFP is expressed as a fusion protein incorporating amino acids 1–39 of σ3 protein at the N terminus. Recombinant virus rsT3D/S4-GFP was recovered following plasmid transfection of L cells stably expressing WT σ3 protein, which is required for propagation of this strain (Figure S1). RT-PCR analysis using primers specific for T3D S4 and GFP confirmed incorporation of a recombinant S4-GFP gene segment into rsT3D/S4-GFP (Figure 7B). Infection of L cells with rsT3D/S4-GFP resulted in expression of GFP and viral inclusion-forming proteins μNS and σNS but not σ3 (Figures 7C and 7D). The capacity of rsT3D/S4-GFP to express GFP was not altered through four passages (data not shown). These results demonstrate that reovirus can be engineered to express foreign genes.
Figure 7.
Expression of GFP by rsT3D/S4-GFP
(A) Schematic of pT7-S4GFP. The GFP ORF is flanked by S4 gene nucleotides 1–149 and 769–1196.
(B) RT-PCR analysis of rsT3D and rsT3D/S4-GFP. Viral dsRNA was extracted from purified virions and subjected to RT-PCR using primers specific for T3D S4 and GFP sequences. Numbers delineate the S4 RNA nucleotide position corresponding to the 5′ end of S4-specific primers.
(C) Viral protein expression in cells infected with rsT3D/S4-GFP. L cells were infected with rsT3D or rsT3D/S4-GFP at an MOI of 1 PFU per cell and incubated for 24 hr. Cell lysates were analyzed by immunoblotting using antibodies specific for μNS, σNS, and σ3 proteins.
(D) Image analysis of cells infected with rsT3D/S4-GFP. L cells were infected with rsT3D/S4-GFP, stained with antibodies specific for μNS (blue) and σ3 (red) proteins at 24 hr postinfection, and imaged by confocal laser scanning microscopy. Scale bar, 10 μM.
Discussion
The absence of DNA intermediates in the life cycle of RNA viruses poses a technical challenge to genetic analysis of viral phenotypes. Prior to the development of reverse genetics, or “marker rescue,” for RNA viruses of animals, in which plasmid-borne cDNAs of viral genomes initiate synthesis of replication-competent RNAs, classical Darwinian methods were used to select viral mutants that could be subjected to correlative genetic studies—so-called “forward genetics.” However, reverse genetics technology permits testing of tightly focused, rational hypotheses about complex questions of virus structure, virus-cell interactions, and viral pathogenesis through direct engineering of the viral genome without a need to devise complicated selection strategies for isolation of viral mutants. Furthermore, reverse genetics of RNA viruses has supported rapid generation of vaccines against these and other infectious agents and propelled their use as gene delivery vehicles (Blaney et al., 2006, Horimoto and Kawaoka, 2006, Riezebos-Brilman et al., 2006).
We developed a fully plasmid-based reverse genetics technology for mammalian reoviruses. This system permits selective introduction of desired mutations into cloned cDNAs encoding each of the ten viral gene segments, followed by isolation of mutant viruses from cells transfected with the plasmid constructs. Moreover, gene segment cDNAs can be manipulated to facilitate expression of a transgene. Importantly, recombinant viruses are generated without a requirement for helper virus and free of any selection. Thus, this new technology provides a means to directly and precisely engineer the viral genome in the context of infectious virus.
We used the newly developed plasmid-based reovirus reverse genetics system to engineer mutations in the σ1 and σ3 proteins. These proteins form part of the viral outer capsid, which is responsible for numerous major events in reovirus interaction with the cell and host, including attachment, disassembly within endosomes, penetration of cell membranes, induction of apoptosis, growth in the intestine, pathways of spread, neurovirulence, and tropism within the CNS (for reviews, see Chandran and Nibert, 2003, O'Donnell et al., 2003, Guglielmi et al., 2006). Therefore, we initially applied reverse genetics technology to the study of outer-capsid proteins to better understand how these proteins mediate critical steps in reovirus replication and disease.
The infectivity of ISVPs of reovirus strain T1L in L cells is approximately 10-fold greater than that of T3D ISVPs (Nibert et al., 1995). This difference in infectivity is hypothesized to be a direct result of σ1 cleavage (Nibert et al., 1995, Chappell et al., 1998), presumably due to removal of the JAM-A-binding region of the molecule. Although the T249I substitution in expressed T3D σ1 renders it resistant to cleavage by trypsin (Chappell et al., 1998), until now it has not been possible to define the role of σ1 cleavage in T3D infectivity for lack of means to generate a mutant T3D virus with the T249I change. This virus has been generated using reverse genetics, and our findings indicate that cleavage susceptibility of viral attachment protein σ1 due to a single polymorphism at amino acid position 249 is the basis for reduced infectivity of T3D ISVPs relative to virions (Figure 4C) and contributes to diminished growth of T3D in the murine intestine following PO inoculation (Figure 5A).
Previous studies of T3D-derived reovirus strains with altered disassembly kinetics point to a critical role for sequences in the virion-distal, C-terminal lobe of σ3 in susceptibility to acid-dependent proteolysis. A C-terminal Y354H mutation in σ3 protein of strain T3D was selected during persistent infection of L cells (PI viruses) (Wetzel et al., 1997) and by serial passage of virus in L cells treated with either E64 (D-EA viruses) (Ebert et al., 2001) or ammonium chloride (ACA-D viruses) (Clark et al., 2006). Using reovirus reverse genetics, the Y354H substitution was introduced into a WT T3D genetic background, and the resultant virus, rsT3D-σ3Y354H, demonstrated accelerated kinetics of σ3 cleavage (Figure 6B) and diminished sensitivity to the growth-inhibitory effects of E64 (Figure 6C). Residue 354 is located in a position thought to be important for controlling access to protease-hypersensitive regions in σ3, residues 208–214 and 238–242, thereby influencing σ3 cleavage kinetics (Jané-Valbuena et al., 2002). Therefore, it appears that residue 354 in σ3 is a gatekeeper for the viral outer capsid, serving to regulate the balance between viral stability and an irreversible, proteolytically triggered disassembly cascade committing the virion particle to either replication or inactivation.
We exploited the reovirus reverse genetics system to develop a gene-delivery vehicle by replacing the σ3 ORF in the S4 plasmid with a GFP-encoding cDNA (Figure 7). The resultant virus, rsT3D/S4-GFP, expresses GFP through successive passages in cell culture. These results reveal the potential for exploitation of reovirus as a gene-transduction vector with application in the development of new mucosal vaccines, more effective oncolytic agents (Coffey et al., 1998), and high-level expression of foreign genes in animal cells. Ideal reovirus vectors will contain stable σ1 proteins and combine excellent extracellular stability with highly efficient intracellular disassembly. We find that each of these parameters can be independently adjusted through strategic alterations in outer-capsid proteins. Manipulation of inner-capsid proteins and the genomic RNA itself should allow construction of viruses able to circumvent other aspects of virus-cell and virus-host interactions that pose potential barriers to antigen and gene delivery by reovirus.
Our results indicate that productive viral infection is established in a small fraction of L cells, approximately 1 in 105–106 cells, transfected with plasmids encoding the ten reovirus gene segments (Figure 1D). Thus, the reovirus plasmid-based marker rescue system is suited to the isolation of viable viral clones by plaque assay, followed by expansion in cell culture to attain quantities of virus sufficient for phenotypic analyses. Manipulations that severely cripple viral replication or particle assembly are more challenging to study because these changes may prohibit virus isolation. However, recovery of a GFP-expressing virus, rsT3D/S4-GFP, demonstrates that marker rescue of lethal mutations is possible by transcomplementation (in this case with WT σ3 protein) (Figure 7 and Figure S1). This result also agrees with our previous finding that inhibition of reovirus replication by RNAi-mediated reduction of viral protein synthesis is reversible by transcomplementation with ectopically expressed WT protein (Kobayashi et al., 2006). Furthermore, transcomplementation allows precise definition through systematic mutational analysis of functional domains in reovirus proteins that are essential for viral replication (Kobayashi et al., 2006). It should be possible to apply this technique to the new marker rescue system for delineation of structure-function relationships in reovirus proteins and RNA.
Quantitative success of plasmid-initiated reovirus infection in this reverse genetics system probably is not limited by the amount of template or transfection efficiency, since the molar ratio of plasmid to target cell is approximately 5 × 104, and increasing the amount of plasmid does not effect higher viral yields from cotransfection lysates (data not shown). Furthermore, it does not appear that infection efficiency is limited by the absence of viral replication proteins during early steps of replication because high-level expression of the replication proteins μ2, μNS, and σNS, which collaborate to form viral inclusions in infected cells (Mbisa et al., 2000, Broering et al., 2002, Becker et al., 2003, Miller et al., 2003), did not enhance viral recovery (data not shown). Perhaps the presence of other viral or cellular factors associated with natural infection by intact virion particles is required for maximal reovirus infectivity.
Presently, no entirely plasmid-based reverse genetics system has been described for any other dsRNA virus of animals. Although each genus within this constellation of viruses bears unique biologic characteristics and physiochemical properties, there are nonetheless numerous unifying features of virion particle structure and replication mechanisms that should allow principles and techniques established in this study to be applied broadly across the group. We expect that new insights into mammalian reovirus replication learned with the use of this reverse genetics system will be quickly extrapolated to other Reoviridae family members, leading to accelerated development of analogous marker-rescue technologies for those viruses.
Experimental Procedures
Cells and Viruses
L cells and HeLa cells were maintained as described (Barton et al., 2001a). Reovirus strains T1L and T3D are laboratory stocks originally obtained from Dr. Bernard Fields. Virus was purified after growth in L cells by CsCl-gradient centrifugation (Furlong et al., 1988). Purified 35S-methionine-labeled virions were prepared as described (Nibert et al., 1995). Attenuated vaccinia virus strain rDIs-T7pol expressing T7 RNA polymerase was propagated in chick embryo fibroblasts as described (Ishii et al., 2002).
Plasmid Construction
Full-length reovirus cDNAs were amplified by RT-PCR using viral dsRNA extracted from purified virions as template. Amplified cDNAs were initially cloned into pBluescript II SK (−) (Stratagene) for the T3D L1, L2, and L3 genes or pCR 2.1 (Invitrogen) for the T3D M1, M2, M3, S1, S2, S3, and S4 genes and the T1L S1 gene (Table S3). To generate pT7-L1T3D, pT7-L2T3D, pT7-L3T3D, pT7-M1T3D, pT7-M2T3D, pT7-M3T3D, pT7-S2T3D, pT7-S3T3D, and pT7-S4T3D, viral cDNA-containing fragments were subcloned into p3E5EGFP (Watanabe et al., 2004). Viral cDNAs fused at their native 5′ termini to the phage T7 RNA polymerase promoter were inserted into p3E5EGFP by partial or complete replacement of plasmid sequences encoding GFP and the Ebola virus leader and trailer, resulting in ligation of native 3′ termini to the HDV ribozyme sequence. To generate pBacT7-S1T3D and pBacT7-S1T1L, encoding the T3D S1 and T1L S1 genes, respectively, S1 cDNAs fused to the T7 promoter and a portion of the HDV ribozyme were first cloned into the BseRI site of p3E5EGFP, and fragments containing the S1 gene flanked 5′ by the T7 promoter and 3′ by the HDV ribozyme and T7 terminator sequences were subcloned into the XbaI site of pBacPAK8 (Clontech). pBacT7-S1T3D and pT7-S4T3D were used as templates to generate mutant constructs pBacT7-S1T3DT249I and pT7-S4T3DY354H, respectively, by introduction of specific nucleotide substitutions using the QuickChange site-directed mutagenesis kit (Stratagene) (Table S4). To generate pT7-S4GFP, S4 nucleotide sequences 150–768 within pT7-S4T3D were replaced with the GFP ORF. Nucleotide sequences of recombinant plasmids were confirmed by DNA sequencing. Detailed description of cloning strategies is provided in the Supplemental Data.
Plasmid Transfection and Recovery of Recombinant Virus
Monolayers of L cells at approximately 90% confluence (3 × 106 cells) in 60 mm dishes (Costar) were infected with rDIs-T7pol at an MOI of ∼0.5 TCID50. At 1 hr postinfection, cells were cotransfected with ten plasmid constructs representing the cloned T3D genome—pT7-L1T3D (2 μg), pT7-L2T3D (2 μg), pT7-L3T3D (2 μg), pT7-M1T3D (1.75 μg), pT7-M2T3D (1.75 μg), pT7-M3T3D (1.75 μg), pBacT7-S1T3D (2 μg), pT7-S2T3D (1.5 μg), pT7-S3T3D (1.5 μg), and pT7-S4T3D (1.5 μg)—using 3 μl of TransIT-LT1 transfection reagent (Mirus) per microgram of plasmid DNA. Following 0–5 days of incubation, recombinant virus was isolated from transfected cells by plaque purification on monolayers of L cells (Virgin et al., 1988). Electrophoretic analysis of viral dsRNA was performed as described (Wilson et al., 1996). Confirmation of mutations in the S1, S4, and L1 genes of recombinant viruses was performed using the Onestep RT-PCR kit (Qiagen), gene-specific primer sets, and viral dsRNA extracted from purified virions as template. PCR products were analyzed following electrophoresis in Tris-borate-EDTA agarose gels or purified and subjected directly to sequence analysis.
Infectious Center and Viral Yield Assays
L cells were cotransfected with ten plasmids corresponding to the T3D genome as described. For infectious center assays, transfected cells were released from plates by trypsin treatment at various intervals posttransfection, washed, counted, diluted, and applied directly to monolayers of untreated L cells (Dermody et al., 1995), which were processed for plaque assay (Virgin et al., 1988). For viral yield assays, transfected L cells were lysed by freezing and thawing, and viral titers in cell lysates were determined by plaque assay (Virgin et al., 1988).
Immunofluorescence Detection of Reovirus Infection
Parental L cells or L cell transfectants selected for stable expression of σ3 protein (5 × 104) were plated on glass coverslips in 24-well plates (Costar) and infected at an MOI of 10,000 (T3D and rsT3D) or 20,000 (rsT3D/S4-GFP) particles per cell. Following incubation at 37°C for various intervals, cells were fixed and stained for μNS and σ3 proteins as described (Maginnis et al., 2006). Images were acquired using a Zeiss LSM 510 META inverted confocal system (Carl Zeiss) with a Zeiss inverted Axiovert 200M microscope and a Plan-APOCHROMAT 63×/1.4 NA oil immersion DIC objective. Images were processed using MetaMorph image analysis software (Molecular Devices).
Infectivity of Recombinant Viruses
Monolayers of L or L-σ3 cells (2.5 to 5 × 105) in 24-well plates or suspension cultures of MEL cells (5 × 105 cells/ml) were infected with virus at an MOI of 1–2 PFU/cell. After 1 hr adsorption at room temperature, the inoculum was removed, cells were washed twice with PBS, and fresh medium was added. Cells were incubated at 37°C for various intervals, and viral titers in cell lysates were determined by plaque assay (Virgin et al., 1988).
Analysis of Viral Capsid Proteins after Protease Treatment
Purified virions at a concentration of either 2 × 1012 particles/ml (trypsin) or 9 × 1012 particles/ml (chymotrypsin) were digested with either 50 μg/ml of N α-p-tosyl-L-sulfonyl phenylalanyl chloromethyl ketone-treated bovine trypsin (Sigma) or 200 μg/ml of Nα-p-tosyl-L-lysine chloromethyl ketone-treated bovine α-chymotrypsin (Sigma) for various intervals at either 25°C (trypsin) or 8°C (chymotrypsin) as described (Nibert et al., 1995, Wetzel et al., 1997). Protease digestion was stopped by adding either 0.5 mg/ml soybean trypsin inhibitor (trypsin) (Sigma) or 5 mM phenylmethyl-sulfonyl fluoride (chymotrypsin) (Sigma) to the treatment mixtures and cooling at 0°C. Viral proteins were resolved by SDS-PAGE and visualized by either autoradiography (Nibert et al., 1995) or staining with Coomassie blue (Wetzel et al., 1997).
Infection of Mice
Newborn C57/BL6 mice weighing 2.0–2.5 grams (2–4 days old) were inoculated perorally or intracranially with 103 or 102 PFU, respectively, of purified reovirus virions diluted in PBS. PO inoculations (50 μl) were delivered intragastrically as described (Rubin and Fields, 1980). IC inoculations (5 μl) were delivered to the left cerebral hemisphere using a Hamilton syringe and 30-gauge needle (Tyler et al., 1985). At various intervals following inoculation, mice were euthanized, and organs were harvested into 1 ml of PBS and homogenized by freezing, thawing, and sonication. Viral titers in organ homogenates were determined by plaque assay (Virgin et al., 1988). Animal husbandry and experimental procedures were performed in accordance with Public Health Service policy and approved by the Vanderbilt University School of Medicine Institutional Animal Care and Use Committee.
Growth of Virus in Cells Treated with E64
Monolayers of L cells (2 × 105) in 24-well plates were preincubated in medium supplemented with 0–200 μM E64 (Sigma) for 4 hr. The medium was removed, and cells were adsorbed with virus at an MOI of 2 PFU/cell. After incubation at 4°C for 1 hr, the inoculum was removed, cells were washed with PBS, and 1 ml of fresh medium supplemented with 0 to 200 μM E64 was added. Cells were incubated at 37°C for 24 hr and frozen and thawed twice. Viral titers in cell lysates were determined by plaque assay (Virgin et al., 1988).
Generation of σ3-Expressing Cells
L cells stably expressing σ3 protein (L-σ3 cells) were selected by transfection of cells with pCXN-S4T3D, which encodes the entire T3D σ3 ORF, and incubation in the presence of 1 mg/ml of geneticin (Invitrogen).
Acknowledgments
We thank Erik Barton, Craig Forrest, and Tim Peters for review of the manuscript and Dirk Reiter, Johannes Schilling, and Thilo Stehle for assistance in preparation of the figures. We thank Yoshihiro Kawaoka for plasmid p3E5EGFP and Tatsuo Miyamura and Koji Ishii for vaccinia virus rDIs-T7pol. This research was supported by a fellowship from the Naito Foundation (T.K.), Public Health Service awards T32 GM07347 (A.A.R.A. and E.A.E.), T32 CA09385 (K.W.B.), T32 GM08554 (K.M.G.), T32 AI49824 and F32 AI71440 (G.H.H.), T32 AI07611 (E.M.J.), T32 AI07281 (M.S.M.), K08 AI62862 (J.D.C.), R01 AI32539, and R37 AI38296, and the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service awards P30 CA68485 for the Vanderbilt-Ingram Cancer Center and P60 DK20593 for the Vanderbilt Diabetes Research and Training Center.
Published: April 18, 2007
Footnotes
The Supplemental Data include Supplemental Experimental Procedures, four supplemental tables, and one supplemental figure and can be found with this article online at http://www.cellhostandmicrobe.com/cgi/content/full/1/2/147/DC1/.
Contributor Information
James D. Chappell, Email: jim.chappell@vanderbilt.edu.
Terence S. Dermody, Email: terry.dermody@vanderbilt.edu.
Accession Numbers
GenBank accession numbers for cDNAs corresponding to the T3D L1, L2, L3, M1, M2, M3, S1, S2, S3, and S4 genes and the T1L S1 gene are provided in Table S3.
Supplemental Data
References
- Baer G.S., Dermody T.S. Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 1997;71:4921–4928. doi: 10.1128/jvi.71.7.4921-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton E.S., Connolly J.L., Forrest J.C., Chappell J.D., Dermody T.S. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J. Biol. Chem. 2001;276:2200–2211. doi: 10.1074/jbc.M004680200. [DOI] [PubMed] [Google Scholar]
- Barton E.S., Forrest J.C., Connolly J.L., Chappell J.D., Liu Y., Schnell F., Nusrat A., Parkos C.A., Dermody T.S. Junction adhesion molecule is a receptor for reovirus. Cell. 2001;104:441–451. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- Becker M.M., Peters T.R., Dermody T.S. Reovirus σNS and μNS proteins form cytoplasmic inclusion structures in the absence of viral infection. J. Virol. 2003;77:5948–5963. doi: 10.1128/JVI.77.10.5948-5963.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaney J.E., Jr., Durbin A.P., Murphy B.R., Whitehead S.S. Development of a live attenuated dengue virus vaccine using reverse genetics. Viral Immunol. 2006;19:10–32. doi: 10.1089/vim.2006.19.10. [DOI] [PubMed] [Google Scholar]
- Bodkin D.K., Fields B.N. Growth and survival of reovirus in intestinal tissue: Role of the L2 and S1 genes. J. Virol. 1989;63:1188–1193. doi: 10.1128/jvi.63.3.1188-1193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodkin D.K., Nibert M.L., Fields B.N. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J. Virol. 1989;63:4676–4681. doi: 10.1128/jvi.63.11.4676-4681.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsa J., Sargent M.D., Lievaart P.A., Copps T.P. Reovirus: Evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology. 1981;111:191–200. doi: 10.1016/0042-6822(81)90664-4. [DOI] [PubMed] [Google Scholar]
- Broering T.J., Parker J.S., Joyce P.L., Kim J., Nibert M.L. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 2002;76:8285–8297. doi: 10.1128/JVI.76.16.8285-8297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.A., Shelling P., Wetzel J.D., Johnson E.M., Wilson G.A.R., Forrest J.C., Aurrand-Lions M., Imhof B., Stehle T., Dermody T.S. Junctional adhesion molecule-A serves as a receptor for prototype and field-isolate strains of mammalian reovirus. J. Virol. 2005;79:7967–7978. doi: 10.1128/JVI.79.13.7967-7978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K., Nibert M.L. Animal cell invasion by a large nonenveloped virus: Reovirus delivers the goods. Trends Microbiol. 2003;11:374–382. doi: 10.1016/s0966-842x(03)00178-1. [DOI] [PubMed] [Google Scholar]
- Chandran K., Zhang X., Olson N.H., Walker S.B., Chappell J.D., Dermody T.S., Baker T.S., Nibert M.L. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J. Virol. 2001;75:5335–5342. doi: 10.1128/JVI.75.11.5335-5342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K., Farsetta D.L., Nibert M.L. Strategy for nonenveloped virus entry: A hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J. Virol. 2002;76:9920–9933. doi: 10.1128/JVI.76.19.9920-9933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.D., Gunn V.L., Wetzel J.D., Baer G.S., Dermody T.S. Mutations in type 3 reovirus that determine binding to sialic acid are contained in the fibrous tail domain of viral attachment protein σ1. J. Virol. 1997;71:1834–1841. doi: 10.1128/jvi.71.3.1834-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.D., Barton E.S., Smith T.H., Baer G.S., Duong D.T., Nibert M.L., Dermody T.S. Cleavage susceptibility of reovirus attachment protein σ1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the σ1 neck. J. Virol. 1998;72:8205–8213. doi: 10.1128/jvi.72.10.8205-8213.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.D., Duong J.L., Wright B.W., Dermody T.S. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J. Virol. 2000;74:8472–8479. doi: 10.1128/jvi.74.18.8472-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J.D., Prota A., Dermody T.S., Stehle T. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 2002;21:1–11. doi: 10.1093/emboj/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.M., Wetzel J.D., Bayley J., Ebert D.H., McAbee S.A., Stoneman E.K., Baer G.S., Zhu Y., Wilson G.J., Prasad B.V.V., Dermody T.S. Reovirus variants selected for resistance to ammonium chloride have mutations in viral outer-capsid protein σ3. J. Virol. 2006;80:671–681. doi: 10.1128/JVI.80.2.671-681.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey M.C., Strong J.E., Forsyth P.A., Lee P.W. Reovirus therapy of tumors with activated Ras pathway. Science. 1998;282:1332–1334. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- Dermody T.S., Chappell J.D., Hofler J.G., Kramp W., Tyler K.L. Eradication of persistent reovirus infection from a B-cell hybridoma. Virology. 1995;212:272–276. doi: 10.1006/viro.1995.1483. [DOI] [PubMed] [Google Scholar]
- Ebert D.H., Wetzel J.D., Brumbaugh D.E., Chance S.R., Stobie L.E., Baer G.S., Dermody T.S. Adaptation of reovirus to growth in the presence of protease inhibitor E64 segregates with a mutation in the carboxy terminus of viral outer-capsid protein σ3. J. Virol. 2001;75:3197–3206. doi: 10.1128/JVI.75.7.3197-3206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D.H., Deussing J., Peters C., Dermody T.S. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 2002;277:24609–24617. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M.L., Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Fields B.N. Temperature-sensitive mutants of reovirus type 3 features of genetic recombination. Virology. 1971;46:142–148. doi: 10.1016/0042-6822(71)90013-4. [DOI] [PubMed] [Google Scholar]
- Furlong D.B., Nibert M.L., Fields B.N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y., Morgan M., Muthukrishnan S., Shatkin A.J. Reovirus messenger RNA contains a methylated blocked 5′-terminal structure M7G(5′)ppp(5′)GmpCp- Proc. Natl. Acad. Sci. USA. 1975;72:362–366. doi: 10.1073/pnas.72.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y., Muthukrishnan S., Shatkin A.J. 5′-Terminal M7G(5′)ppp(5′)Gmp in vivo: Identification in reovirus genome RNA. Proc. Natl. Acad. Sci. USA. 1975;72:742–745. doi: 10.1073/pnas.72.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi K.M., Johnson E.M., Stehle T., Dermody T.S. Attachment and cell entry of mammalian orthoreovirus. Curr. Top. Microbiol. Immunol. 2006;309:1–38. doi: 10.1007/3-540-30773-7_1. [DOI] [PubMed] [Google Scholar]
- Horimoto T., Kawaoka Y. Strategies for developing vaccines against H5N1 influenza A viruses. Trends Mol. Med. 2006;12:506–514. doi: 10.1016/j.molmed.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Ishii K., Ueda Y., Matsuo K., Matsuura Y., Kitamura T., Kato K., Izumi Y., Someya K., Ohsu T., Honda M., Miyamura T. Structural analysis of vaccinia virus DIs strain: Application as a new replication-deficient viral vector. Virology. 2002;302:433–444. doi: 10.1006/viro.2002.1622. [DOI] [PubMed] [Google Scholar]
- Jané-Valbuena J., Breun L.A., Schiff L.A., Nibert M.L. Sites and determinants of early cleavages in the proteolytic processing pathway of reovirus surface protein σ3. J. Virol. 2002;76:5184–5197. doi: 10.1128/JVI.76.10.5184-5197.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapikian A., Hoshino Y., Chanock R. Rotaviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott-Raven; Philadelphia: 2001. pp. 1787–1833. [Google Scholar]
- Kobayashi T., Chappell J.D., Danthi P., Dermody T.S. Gene-specific inhibition of reovirus replication by RNA interference. J. Virol. 2006;80:9053–9063. doi: 10.1128/JVI.00276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoto S., Sasaki J., Taniguchi K. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. USA. 2006;103:4646–4651. doi: 10.1073/pnas.0509385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maginnis M.S., Forrest J.C., Kopecky-Bromberg S.A., Dickeson S.K., Santoro S.A., Zutter M.M., Nemerow G.R., Bergelson J.M., Dermody T.S. β1 integrin mediates internalization of mammalian reovirus. J. Virol. 2006;80:2760–2770. doi: 10.1128/JVI.80.6.2760-2770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Padura I., Lostaglio S., Schneemann M., Williams L., Romano M., Fruscella P., Panzeri C., Stoppacciaro A., Ruco L., Villa A. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbisa J.L., Becker M.M., Zou S., Dermody T.S., Brown E.G. Reovirus μ2 protein determines strain-specific differences in the rate of viral inclusion formation in L929 cells. Virology. 2000;272:16–26. doi: 10.1006/viro.2000.0362. [DOI] [PubMed] [Google Scholar]
- Miller C.L., Broering T.J., Parker J.S., Arnold M.M., Nibert M.L. Reovirus σNS protein localizes to inclusions through an association requiring the μNS amino terminus. J. Virol. 2003;77:4566–4576. doi: 10.1128/JVI.77.8.4566-4576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert M.L., Chappell J.D., Dermody T.S. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved σ1 protein. J. Virol. 1995;69:5057–5067. doi: 10.1128/jvi.69.8.5057-5067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard A.L., Chandran K., Zhang X., Parker J.S., Baker T.S., Nibert M.L. Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 2004;78:8732–8745. doi: 10.1128/JVI.78.16.8732-8745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell S.M., Hansberger M.W., Dermody T.S. Viral and cellular determinants of apoptosis induced by mammalian reovirus. Int. Rev. Immunol. 2003;22:477–503. doi: 10.1080/08830180305212. [DOI] [PubMed] [Google Scholar]
- Olland A.M., Jané-Valbuena J., Schiff L.A., Nibert M.L., Harrison S.C. Structure of the reovirus outer capsid and dsRNA-binding protein σ3 at 1.8 Å resolution. EMBO J. 2001;20:979–989. doi: 10.1093/emboj/20.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.S., Broering T.J., Kim J., Higgins D.E., Nibert M.L. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 2002;76:4483–4496. doi: 10.1128/JVI.76.9.4483-4496.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezebos-Brilman A., de Mare A., Bungener L., Huckriede A., Wilschut J., Daemen T. Recombinant alphaviruses as vectors for anti-tumour and anti-microbial immunotherapy. J. Clin. Virol. 2006;35:233–243. doi: 10.1016/j.jcv.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Roner M.R., Joklik W.K. Reovirus reverse genetics: Incorporation of the CAT gene into the reovirus genome. Proc. Natl. Acad. Sci. USA. 2001;98:8036–8041. doi: 10.1073/pnas.131203198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roner M.R., Steele B.G. Localizing the reovirus packaging signals using an engineered m1 and s2 ssRNA. Virology. 2007;358:89–97. doi: 10.1016/j.virol.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Roner M.R., Nepliouev I., Sherry B., Joklik W.K. Construction and characterization of a reovirus double temperature-sensitive mutant. Proc. Natl. Acad. Sci. USA. 1997;94:6826–6830. doi: 10.1073/pnas.94.13.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D.H., Fields B.N. Molecular basis of reovirus virulence: Role of the M2 gene. J. Exp. Med. 1980;152:853–868. doi: 10.1084/jem.152.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D.H., Wetzel J.D., Williams W.V., Cohen J.A., Dworkin C., Dermody T.S. Binding of type 3 reovirus by a domain of the σ1 protein important for hemagglutination leads to infection of murine erythroleukemia cells. J. Clin. Invest. 1992;90:2536–2542. doi: 10.1172/JCI116147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein S.C., Schur P.H. Immunofluorescent localization of double-stranded RNA in reovirus-infected cells. Virology. 1970;41:564–566. doi: 10.1016/0042-6822(70)90178-9. [DOI] [PubMed] [Google Scholar]
- Tyler K.L. Mammalian reoviruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams & Wilkins; Philadelphia: 2001. pp. 1729–1745. [Google Scholar]
- Tyler K.L., Bronson R.T., Byers K.B., Fields B.N. Molecular basis of viral neurotropism: Experimental reovirus infection. Neurology. 1985;35:88–92. doi: 10.1212/wnl.35.1.88. [DOI] [PubMed] [Google Scholar]
- Tyler K.L., Barton E.S., Ibach M.L., Robinson C., Valyi-Nagy T., Campbell J.A., Clarke P., O'Donnell S.M., Wetzel J.D., Dermody T.S. Isolation and molecular characterization of a novel type 3 reovirus from a child with meningitis. J. Infect. Dis. 2004;189:1664–1675. doi: 10.1086/383129. [DOI] [PubMed] [Google Scholar]
- Virgin H.W., IV, Bassel-Duby R., Fields B.N., Tyler K.L. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing) J. Virol. 1988;62:4594–4604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Watanabe T., Noda T., Takada A., Feldmann H., Jasenosky L.D., Kawaoka Y. Production of novel Ebola virus-like particles from cDNAs: An alternative to Ebola virus generation by reverse genetics. J. Virol. 2004;78:999–1005. doi: 10.1128/JVI.78.2.999-1005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel J.D., Wilson G.J., Baer G.S., Dunnigan L.R., Wright J.P., Tang D.S.H., Dermody T.S. Reovirus variants selected during persistent infections of L cells contain mutations in the viral S1 and S4 genes and are altered in viral disassembly. J. Virol. 1997;71:1362–1369. doi: 10.1128/jvi.71.2.1362-1369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener J.R., Joklik W.K. The sequences of the reovirus serotype 1, 2, and 3 L1 genome segments and analysis of the mode of divergence of the reovirus serotypes. Virology. 1989;169:194–203. doi: 10.1016/0042-6822(89)90055-x. [DOI] [PubMed] [Google Scholar]
- Wilson G.J., Wetzel J.D., Puryear W., Bassel-Duby R., Dermody T.S. Persistent reovirus infections of L cells select mutations in viral attachment protein σ1 that alter oligomer stability. J. Virol. 1996;70:6598–6606. doi: 10.1128/jvi.70.10.6598-6606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G.J., Nason E.L., Hardy C.S., Ebert D.H., Wetzel J.D., Prasad B.V.V., Dermody T.S. A single mutation in the carboxy terminus of reovirus outer-capsid protein σ3 confers enhanced kinetics of σ3 proteolysis, resistance to inhibitors of viral disassembly, and alterations in σ3 structure. J. Virol. 2002;76:9832–9843. doi: 10.1128/JVI.76.19.9832-9843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.