Summary
Meiotic development in yeast requires the coordinated induction of transient waves of gene transcription. The present study investigates the regulation of Ume6p, a mitotic repressor of the “early” class of meiosis-specific genes. Western blot analysis revealed that Ume6p is destroyed early in meiosis by Cdc20p, an activator of the anaphase promoting complex/cyclosome (APC/C) ubiquitin ligase. This control appears direct as Cdc20p and Ume6p associate in vivo and APC/CCdc20 ubiquitylates Ume6p in vitro. Inactivating Cdc20p, or stabilizing Ume6p through mutation, prevented meiotic gene transcription and meiotic progression. During mitotic cell division, Ume6p is protected from destruction by Protein Kinase A phosphorylation of Cdc20p. Complete elimination of Ume6p in meiotic cells requires association with the meiotic inducer Ime1p. These results indicate that Ume6p degradation is required for normal meiotic gene induction and meiotic progression. These findings demonstrate a direct connection between the transcription machinery and ubiquitin-mediated proteolysis that is developmentally regulated.
Introduction
Meiosis is the process by which diploid organisms generate haploid gametes capable of sexual reproduction. In the budding yeast Saccharomyces cerevisiae, entry into meiosis is controlled by cell type and nutritional cues. Specifically, only MATa/MATα diploid cells starved for nitrogen and a fermentable carbon source are competent to enter meiosis (see (Kupiec et al., 1997) for review). The nutritional pathway inhibits meiotic induction through Ras-dependent activation of protein kinase A (PKA). Both cell type and nutritional controls converge to regulate Ime1p expression. Ime1p is a master regulator of meiotic induction whose activity most likely represents the first step in initiating this process (Kassir et al., 1988).
Many of the genes required for meiotic events in yeast display a similar expression profile in that their mRNA levels are very low during mitotic cell division then transiently transcribed at defined stages during development. These genes generally cluster into three classes termed early, middle and late based on their timing of expression (Chu et al., 1998) although additional groups are present (Primig et al., 2000). Ume6p is a C6 zinc cluster DNA binding protein that is required for the mitotic repression of several early meiotic genes (e.g., SPO13, IME2) (Bowdish and Mitchell, 1993; Strich et al., 1994). Ume6p represses transcription by recruiting the histone deacetylase Rpd3p through the general co-repressor Sin3p (Kadosh and Struhl, 1998) and the chromatin remodeling factor Isw2p (Goldmark et al., 2000). Although a repressor, mutants lacking Ume6p fail to execute either meiotic division or form spores (Strich et al., 1994). Several reports have suggested a model that Ume6p repression is relieved when it is converted from a repressor to an activator by association with the meiotic inducer Ime1p (Bowdish et al., 1995; Rubin-Bejerano et al., 1996; Washburn and Esposito, 2001),
Similar to the mitotic cell cycle, the transition between one stage of meiosis to the next is controlled by ubiquitin mediated proteolysis (Harper et al., 2002). In budding yeast, the association of one of three highly conserved Trp-Asp (WD) repeat proteins (Ama1p, Cdc20p or Hct1p/Cdh1p) serve as stage-specific activators and specificity factors for the anaphase promoting complex/cyclosome (APC/C) ubiquitin ligase (Cooper et al., 2000; Dawson et al., 1995; Schwab et al., 1997; Sigrist and Lehner, 1997; Visintin et al., 1997). During meiosis, the Cdc20p activated APC/C (APC/CCdc20) promotes chromosome separation through Pds1p destruction (Salah and Nasmyth, 2000) while APC/CAma1 is required for normal exit from meiosis II through destruction of the B-type cyclin Clb1p (Coluccio et al., 2004; Cooper et al., 2000).
In the present study, we report that Ume6p repression is relieved by a two-step proteolysis pathway mediated by APC/CCdc20. Step one induces partial degradation of Ume6p in cultures switched from growth on a fermentable to a non-fermentable carbon source. The second step in which Ume6p is completely eliminated requires meiotic entry and association with Ime1p. These findings provide a direct mechanism for meiotic gene induction and demonstrate a role for APC/CCdc20 in transcriptional control that is independent of its established function in mitotic cell cycle progression.
Results
Ume6p is down regulated early in meiosis
To begin exploring how Ume6p-dependent repression is relieved, we examined Ume6p levels during meiosis. As we were unable to produce high-affinity antibodies against Ume6p, a T7 epitope-tagged derivative of UME6 was generated and used to replace the wild-type gene at its normal chromosomal location under its own promoter (see Experimental procedures). This tagged UME6 allele (T7-UME6) produced a protein that migrated slower during SDS PAGE then would be expected from its predicted 91 kDa size (Fig. 1A) most likely due to the basic nature of the protein (pI 10.4). To test the functionality of T7-Ume6p, a plasmid harboring a spo13-URA3 reporter gene was introduced into this strain. Repression of this reporter gene by T7-Ume6p will make the host strain phenotypically Ura− and resistant to the uracil analog 5-fluoro-orotic acid (Strich et al., 1994). As shown in Fig. 1B, T7-Ume6p functioned similar to the wild-type strain in this assay. In addition, T7-UME6 diploids also execute the meiotic program and form spores with wild-type kinetics (see below). These results indicate that T7-Ume6p functions similar to the wild-type protein with respect to meiotic gene repression and meiotic progression.
Figure 1.
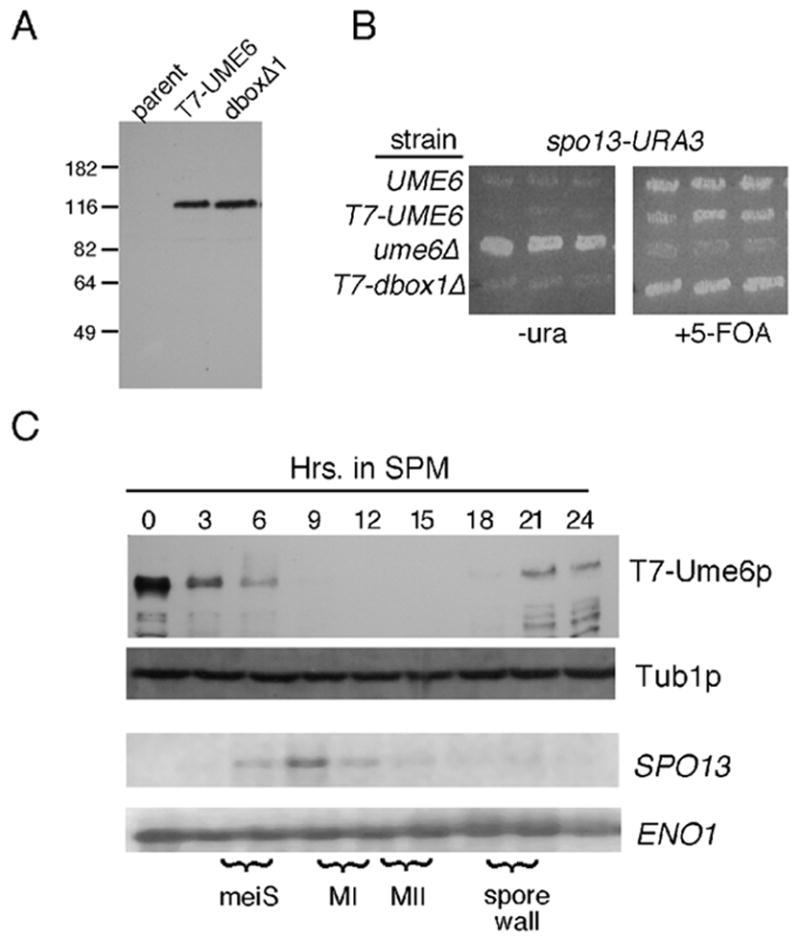
Ume6p regulation during meiosis. Panel A. Vegetative steady state levels of T7-epitope tagged wild-type strain RSY1079 (T7-Ume6p) and destruction box 1 mutant (RSY1180, Ume6pdb1Δ) proteins were determined Western blot analysis. The parental control (RSY335) lacks a T7-tagged UME6 allele. Molecular weight standards (kDa) are given on the left of the panel. Panel B. The indicated strains UME6 (RSY335), T7-UME6 (RSY1079), ume6Δ (RSY853) and T7-dbox1Δ (RSY1080) were transformed with the spo13-URA3 reporter plasmid (pMS49). Three independent transformants were patched on medium lacking uracil (-uracil) or containing the uracil analog 5-FOA. Plates were incubated two days and photographed. Panel C. A meiotic timecourse was conducted with strain RSY1079 and timepoints taken prior (0 hr) and following the shift to sporulation medium (SPM). T7-Ume6p levels were determined by Western blot analysis. Northern blot analysis of SPO13 mRNA was conducted from the same timepoints. The execution of meiotic S phase (meiS), the meiotic divisions (MI, MII) and spore wall formation were determined by FACS analysis, fluorescent microscopy of DAPI stained cells and light microscopy, respectively. Tub1p and ENO1 mRNA levels served as loading controls for the Western and Northern blot analyses, respectively.
To monitor Ume6p levels during meiosis, the T7-UME6 diploid was induced to enter meiosis and timepoints taken. Western blot analysis of total protein preparations revealed that Ume6p levels rapidly declined shortly following transfer to sporulation medium (SPM, Fig. 1C) approximately the time of pre-meiotic S phase (meiS) initiation. Ume6p levels were again detected late in the program after the completion of meiosis I and II during spore wall assembly. Similar results were obtained with different extraction procedures and strain background (data not shown). Interestingly, Northern blot analysis of the same timepoints revealed that SPO13 mRNA levels peaked when Ume6p levels were undetectable (9 hr). These results indicate that Ume6p levels are dynamic during meiosis. As UME6 mRNA levels remain constant during meiosis (Strich et al., 1994), the changes in Ume6p concentrations are most likely due to a post-transcriptional mechanism.
Ume6p degradation requires APC/CCdc20
The reduction in protein levels can occur through increased turnover, decreased translation, or both. To determine if protein turnover was the underlying cause for Ume6p down regulation, we first tested the role of Ama1p, a meiosis-specific Cdc20 family member (Cooper et al., 2000). However, Ume6p levels were reduced in an ama1 mutant with kinetics similar to wild type (data not shown). Similarly, a strain lacking the APC/C activator Hct1p/Cdh1p did not affect Ume6p degradation kinetics (data not shown). The other Cdc20 family member known to have a role in meiosis is Cdc20p itself (Simchen, 1974). Since Cdc20p is required for mitotic cell division (Hartwell et al., 1974), a conditional allele (cdc20-1) was employed. A diploid strain homozygous for the cdc20-1 allele was grown to mid-log phase at the permissive temperature (23°). The culture was transferred to SPM at the permissive temperature for four hrs to allow the cdc20-1 culture to completely exit the cell cycle. After four hours, the cells were shifted to restrictive temperature (34.5°C) and the timecourse continued for 24 hrs. As before, Ume6p levels were rapidly reduced as the culture entered meiosis (Fig. 2A, top panel). However, following the temperature shift, Ume6p levels rebounded to mitotic levels. The delay in Ume6p recovery (6 hr timepoint) was most likely due to the relatively slow equilibration of the large sporulation culture to the restrictive temperature in an air shaker. These results indicate that Cdc20p is required for Ume6p destruction in meiotic cells. An alternative possibility is that Ume6p destruction is delayed due to the meiotic arrest associated with Cdc20-1p inactivation. However, Clb1p, a B-type cyclin that is destroyed by APC/CAma1 (Cooper et al., 2000) is still down regulated in the cdc20-1 strain (Fig. 2A, middle panel) indicating that APC/C-dependent proteolysis is still active. The reduction and delay in Clb1p peak accumulation is due to a reduction in CLB1 transcription (see below).
Figure 2.

Cdc20p is required for Ume6p destruction. Panel A. The cdc20-1 strain RSY809 was transferred to SPM at the permissive temperature (23°) then shifted to the restrictive temperature (34.5°) four hrs later (arrow). T7-Ume6p levels were determined as before. Wild type (RSY335) and cdc20-1 mutant strain (RSY809) harboring CLB1-3HA plasmid pKC426 were treated as just described and Clb1p levels followed by Western blot analysis. Tub1p levels served as loading controls. Panel B. In vitro ubiquitylation assays were conducted with constant Cdc20p concentrations (2.5 μl) for the times indicated (left panel). Increased amounts of Cdc20p (2.5 and 5 μl) were added to the reaction (90 min) with Ume6p or Ume6pdb1Δ as substrate (right panel). The arrow heads indicate unmodified substrate, brackets, ubiquitylated species. The right panel was overexposed compared to the left panel to determine if weak ubiquitylation activity was observed with the destruction box mutant substrate.
To determine if the APC/CCdc20 ligase can directly ubiquitylate Ume6p, an in vitro assay was performed (see Experimental procedures). Incubating Ume6p with APC/CCdc20 produced a modest, time dependent accumulation of higher molecular weight species indicative of poly-ubiquitylation (bracket, Fig. 2B left panel). This modification required an intact destruction box, a degradation signal targeted by APC/CCdc20 (Fig. 2B, right panel). Mutating this destruction box delayed Ume6p destruction in vivo (see below) indicating that it is functional. Taken together, these results indicate that Ume6p is an in vitro substrate of APC/CCdc20.
Cdc20p is required for the normal meiotic gene transcription
Ume6p is the major repressor or early meiotic genes during vegetative growth. Therefore, we next determined whether APC/CCdc20 played a role in controlling meiotic gene transcription. As described above, wild type and cdc20-1 cultures were grown at the permissive temperature (23°) than shifted to the restrictive temperature (34.5°) four hrs following transfer to sporulation medium. A Northern blot of total RNA samples was probed for mRNAs corresponding to members of the early (IME2), early-middle (NDT80) middle (SPS2, SPS4) and late (SPS100) expression classes. In addition, the mRNA levels of IME1, a gene not regulated by Ume6p, were followed to access the ability of each strain to enter meiosis. The expression profiles of IME1 were similar in both strains although peak accumulation was somewhat lower in the mutant (Fig. 3). Interestingly, the induction of IME2 mRNA, a gene repressed by Ume6p, was normal in both strains prior to the shift to restrictive temperature (see 3 hr timepoint). However, following the temperature shift, IME2 mRNA levels were dramatically reduced (approximately 15% of the three hr. timepoint) in the cdc20-1 mutant. In addition, the expression of early-middle and middle gene mRNAs were significantly reduced but still detected. This finding explains the reduced levels of Clb1p protein observed in Figure 2. The transcript levels of the mid-late gene SPS100 were below the limits of detection. As Ime2p is also required for the normal transcriptional induction of later genes (Foiani et al., 1996; Mitchell et al., 1990; Sia and Mitchell, 1995), the middle and late gene transcription defect is likely indirect. These results indicate that Cdc20p is required for the meiotic transcription program.
Figure 3.

Cdc20p is required for normal meiotic gene expression. Wild-type (RSY335) and mutant cdc20-1 (RSY809) strains were induced to enter meiosis at 23°C. After 4 hours in SPM, the cultures were shifted to the restrictive temperature (34.5°C). Total RNA was prepared from samples taken at the times indicated (hrs) following shift to SPM and the transcript levels of the genes indicated were analyzed by Northern blot analysis. ENO1 serves as a loading control. MI and MII indicate the approximate times of meiosis I and meiosis II in the wild-type culture as determined by DAPI analysis.
Ume6p destruction is required for early meiotic gene transcription and meiotic progression
The results just described suggest a correlation between Ume6p destruction and meiotic gene induction. To determine if Ume6p destruction is necessary for meiotic gene expression, a stabilized allele of this factor was sought. Cdc20p recognizes an element termed the destruction box. Ume6p has three canonical destruction box (d-box) motifs (Glotzer et al., 1991) at residues 39, 500, and 573 (see Fig. 4A) and one divergent sequence at amino acid 793 within Ume6p’s zinc-cluster domain. The first destruction box motif was mutated (db1 ) then used to replace the wild-type allele in the chromosome. Ume6pdb1Δ is expressed at levels similar to wild type (Fig. 1A) and represses spo13-URA3 transcription during vegetative growth (Fig. 1B) indicating that this mutant possesses repressor activity. Unlike wild type, Ume6pdb1Δ levels remained elevated for 15 hrs following the shift to SPM (Fig. 4B). Importantly, SPO13 mRNA induction kinetics and the execution of the nuclear divisions were similarly delayed (Fig. 4C). These results indicate that Ume6p destruction is required for the timely induction of EMG as well as execution of meiotic landmark events.
Figure 4.
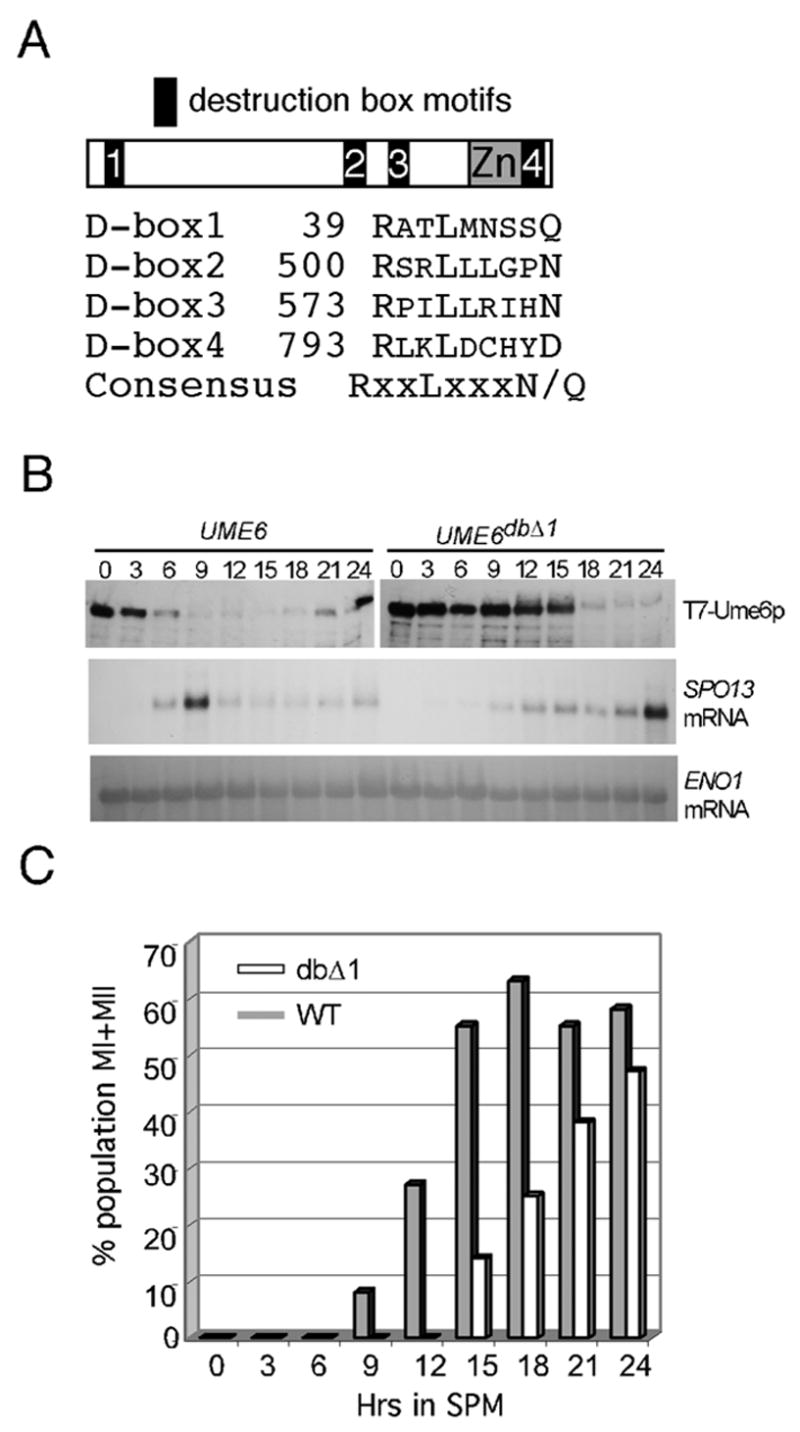
Ume6p destruction is required for normal EMG transcription and meiotic nuclear divisions. Panel A. Schematic of Ume6p indicating location of destruction box motifs and zinc cluster domain (Zn). Sequence alignment of Ume6p destruction box motifs and starting amino acid residue corresponding to the arginine is presented. The consensus sequence is given with X = any amino acid. Panel B. Wild type (RSY1079) and UME6db1Δ (RSY1080) mutant strains were induced to enter meiosis and timepoints taken as indicated (hrs). T7-Ume6p and T7-Ume6pdb1Δ levels were followed by Western blot analysis. SPO13 mRNA levels were monitored by Northern blot analysis. ENO1 mRNA served as the loading control for the Northern analysis. Panel C. The timepoints described in Panel B were examined for the execution of meiosis I and meiosis II by DAPI analysis. The percentage of the population that had undergone one or both divisions in the wild type (shaded bars) or UME6db1Δ strain (open bars) is depicted.
Ume6p is not destroyed during vegetative growth
Ume6p represses EMG transcription in vegetative cells when APC/CCdc20 is active. To determine whether Ume6p is a substrate of APC/CCdc20 during mitotic cell division, T7-Ume6p levels were monitored following a synchronous release from an S-phase arrest by hydroxyurea (HU). Western blot analysis revealed a small (approximately 20%), but reproducible, reduction in Ume6p levels 45 min following HU release (Fig. 5A, upper panel). DAPI staining revealed that anaphase began just prior to the 45 min timepoint and was mostly completed by 60 min. (Fig. 5B). These results indicate that Ume6p is slightly reduced approximately the time of APC/CCdc20 activation. This conclusion is supported by our finding that the destruction box mutant (Ume6pdb1Δ) did not exhibit a similar reduction in levels even though cell cycle progression was identical (Fig. 5A, bottom panel). The modest reduction of Ume6pdb1Δ observed at 75 and 90 min was not reproducible (see wild-type panel). This reduction of Ume6p during mitotic cell division is not, however, sufficient to derepress early meiotic genes (Spellman et al., 1998). These results indicate that Ume6p is partially destroyed by the APC/C during mitotic cell division.
Figure 5.
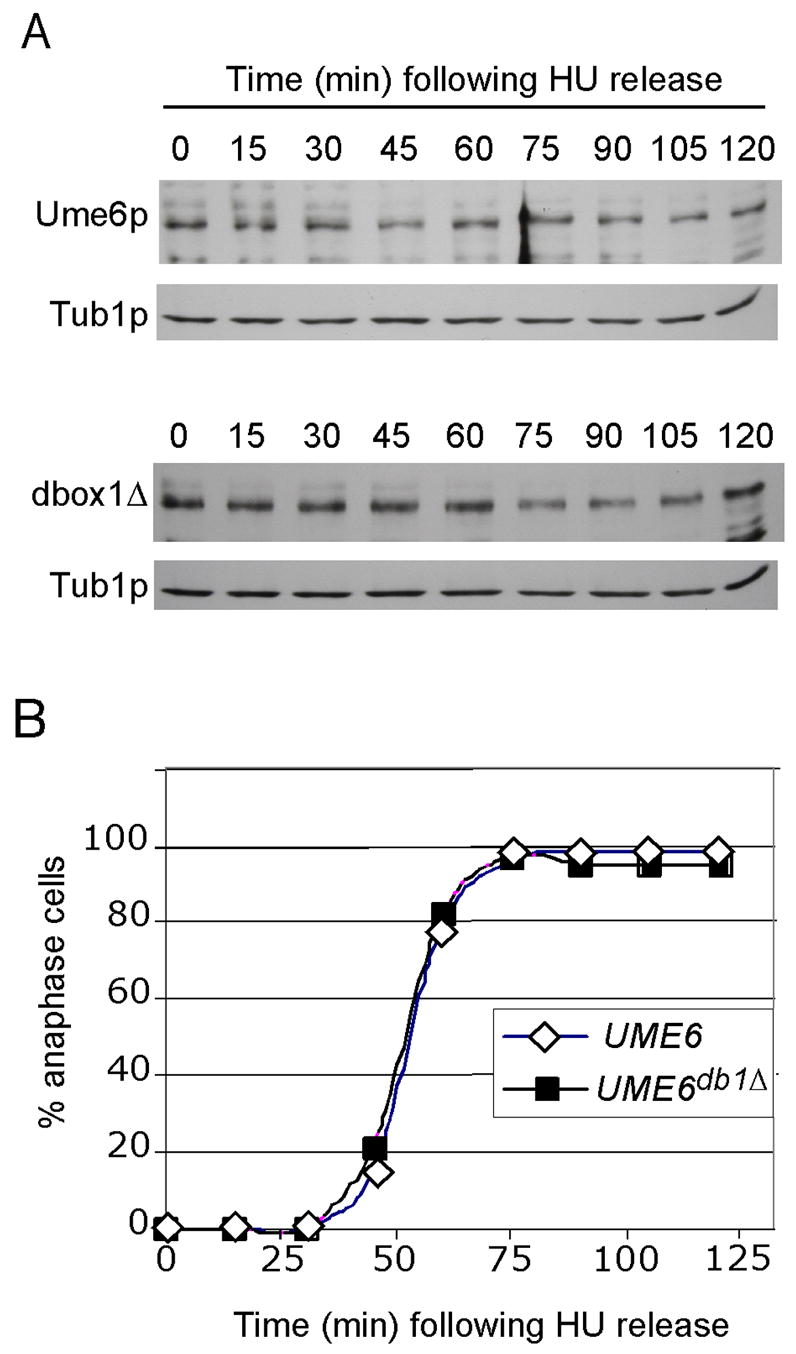
Ume6p is a substrate of APC/CCdc20 during G2-M stage of the cell cycle. Panel A. Haploid strain RSY1069 (T7-UME6, top panel) or RSY1070 (T7-UME6db1Δ) was treated with hydroxyurea (HU) for 2 hrs to arrest the cells in S phase. The cells were washed and placed in fresh medium and timepoints taken as indicated. Ume6p and Ume6pdb1Δ levels were determined by Western blot analysis for every timepoint. Tib1p served as a loading control. Panel B. The execution of anaphase was determined in the samples described in Panel A by monitoring the presence of separated nuclei by DAPI staining and fluorescent microscopy.
Ume6p is protected from destruction in vegetative cells by the RAS/cAMP-PKA pathway
During these studies, we noticed that Ume6p steady state levels were consistently reduced 40–50% in cultures grown in acetate medium versus those utilizing dextrose as the carbon source (Fig. 6A). Moreover, mutating D-box1 suppressed this effect indicating that APC/CCdc20 is also involved in this process. This possibility was verified as inactivating Cdc20p in acetate cultures restored Ume6p to dextrose levels (Fig. 6B). RAS and cAMP dependent Protein Kinase A (PKA) are major regulators of the nutritional response in yeast (Toda et al., 1985). In addition, PKA activity is reduced in cultures grown in acetate versus dextrose medium (Portela et al., 2003). To test whether RAS activity regulates Ume6p stability, we examined the requirement of Cdc25p, the GTP exchange protein that activates Ras1p and Ras2p (Broek et al., 1987). Since Cdc25p is essential for growth, the temperature sensitive allele (cdc25-5) was employed. A T7-UME6 expressing plasmid (pMM285) or vector control was transformed into the cdc25-5 strain. Cultures were grown in dextrose medium at the permissive temperature (23°) to mid-log phase, then split with one half being shifted to the restrictive temperature (37°). After two hrs, the cultures were harvested and protein extracts prepared. Western blot analysis revealed that Ume6p levels were significantly reduced in the cdc25-5 strain following the temperature shift compared to the wild-type control (Fig. 6C). These results indicate that RAS activity is necessary for maintaining normal Ume6p levels during dextrose growth.
Figure 6.
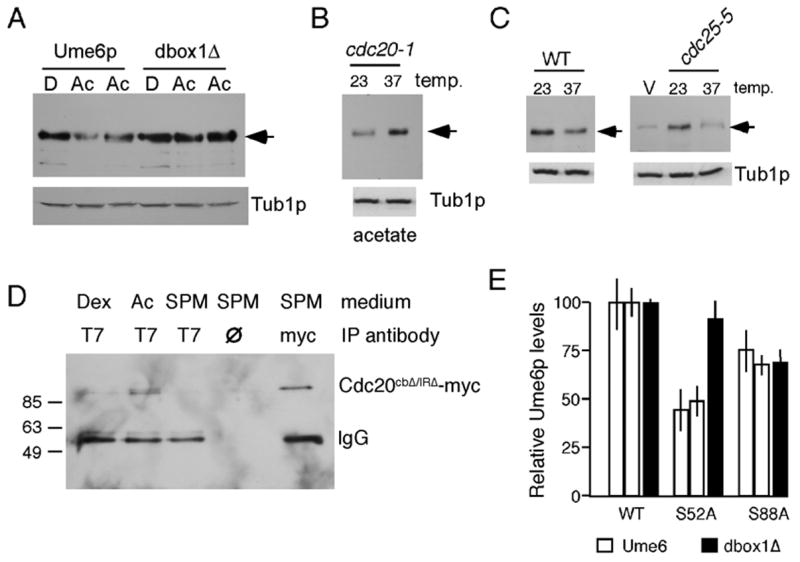
Ume6p levels are regulated by the RAS pathway. Panel A. Wild type (RSY1079) and UME6db1Δ (RSY1080) strains were grown in dextrose (D) or acetate (Ac) medium (two independent cultures) to mid-log phase. Ume6p or Ume6pdb1Δ levels (arrow) were monitored by Western blot analysis. Tub1p served as a loading control. Panel B. T7-Ume6p levels were monitored in the cdc20-1 mutant strain (RSY809) grown to mid-log in acetate medium at permissive (23°), or following two hrs at restrictive (37°), temperature. Panel C. Ume6p levels (arrows) were determined in log phase cultures with the indicated genotypes grown at permissive temperature (23°) and following two hrs at the restrictive (37°) temperature. V = vector control. Panel D. Extracts prepared from log phase RSY1079 cultures in dextrose (Dex), acetate (Ac) or after transfer to sporulation medium (SPM) for 4hr were immunoprecipitated with the indicated antibodies or no antibody (∅). The immunoprecipitates were subjected to Western blot analysis probing for the presence of Cdc20p-13xmyc. Cdc20p-13xmyc migration and expression were verified in the 4 hr sporulation timepoint (right lane). No antibody lane controls for non-specific association of Cdc20p-13xmyc to protein A beads. Molecular weight markers (kDa) are provided on the left. Panel E. Ume6p or Ume6pdb1Δ levels were followed in a cdc20-1 strain (RSY809) harboring wild type (WT), S52A, or S88A CDC20 expression plasmids (see Experimental procedures for details). Error bars indicate standard deviation from the mean.
PKA phosphorylation of Cdc20p protects Ume6p from destruction
A previous report found that Cdc20p was inhibited by PKA phosphorylation on two residues (S52 and S88) following DNA damage (Searle et al., 2004). To determine whether PKA controlled Ume6p levels through Cdc20p, a plasmid harboring either the wild-type CDC20, S52A or S88A allele was introduced into a cdc20-1 strain expressing T7-Ume6p. Two separate experiments, each with two independent transformants, were performed for this study. Cultures were grown to mid-log phase in dextrose medium at the restrictive temperature (37°) to inactivate endogenous Cdc20-1p. Quantitation revealed that steady state T7-Ume6p levels were reduced to approximately 50% in the S52A mutant (P = 0.007, Fig. 6D). This reduction was dependent on Cdc20p as this effect was lost with Ume6pdb1Δ (two independent cultures). The S88A allele also caused a modest, but not statistically significant (P = 0.119) reduction in T7-Ume6p levels. No differences were observed between the wild type and D-box1 mutant in the S88A expressing strain suggesting that the minor reduction observed for Ume6p in this background was most likely indirect. Taken together, these results indicate that Ume6p levels are maintained in dextrose medium by inactivation of Cdc20p by PKA.
Ume6p-Cdc20p association is regulated by carbon source
The Cdc20p protein family functions as adaptors linking the substrate to the catalytically active APC/C ubiquitin ligase (Harper et al., 2002). Therefore, we next tested whether Ume6p-Cdc20p association is regulated by nutritional signals. As Cdc20p interaction triggers Ume6p destruction, a mutant form of Cdc20p was utilized (Cdc20pcbΔ/IRΔ) that lacked two regions (C-box and IR domain) required for APC/C interaction (Burton et al., 2005; Schwab et al., 2001). Previous studies found that overexpression was required to detect Cdc20p containing complexes in co-immunoprecipitation assays (Burton and Solomon, 2001). Co-immunoprecipitation experiments were performed with extracts prepared from a wild-type T7-UME6 diploid harboring a high-copy plasmid expressing 13Xmyc-Cdc20pcbΔ/IRΔ grown in different media. Immunoprecipitates following T7 antibody incubation were subjected to Western blot analysis probing for the presence of Cdc20pcbΔ/IRΔ. Cdc20pcbΔ/IRΔ association with Ume6p was not detected in dextrose medium but was observed in the acetate cultures (Fig. 6E). Cdc20pcbΔ/IRΔ was not observed in the Ume6p immunoprecipitates after four hrs incubation in the sporulation medium (SPM) suggesting either that this interaction is not stable later in development or that Ume6p levels were significantly reduced due to the presence of wild-type Cdc20p. These results indicate that Cdc20p-Ume6p interaction is regulated by carbon source through the RAS-PKA pathway.
Complete Ume6p destruction requires entry into the meiotic program and Ime1p association
During growth in nitrogen rich conditions, Ume6p represses two genes (CAR1, CAR2) involved in nitrogen catabolism (Park et al., 1992; Strich et al., 1994). Therefore, we tested whether Ume6p down regulation was a common response to nitrogen starvation, and not meiotic induction, per se. The T7-UME6 allele was introduced into a diploid strain that was not competent to enter meiosis (MATa/MATa). This strain was grown to mid-log phase in acetate medium and transferred to sporulation medium. Western blot analysis revealed that Ume6p levels were partially reduced by three hrs but then remained constant up to 12 hrs (Fig. 7A). As this strain was growing in acetate medium, this small reduction may be the result of nitrogen starvation. This reduction in Ume6p levels requires APC/CCdc20 activity as Ume6pdb1Δ levels were not affected by exposure to SPM (see Fig. 4C). In addition, complete Ume6p destruction also requires the meiotic inducer Ime1p (Fig. 7B). These results indicate that Ume6p destruction requires meiotic induction and is not simply a response to starvation.
Figure 7.
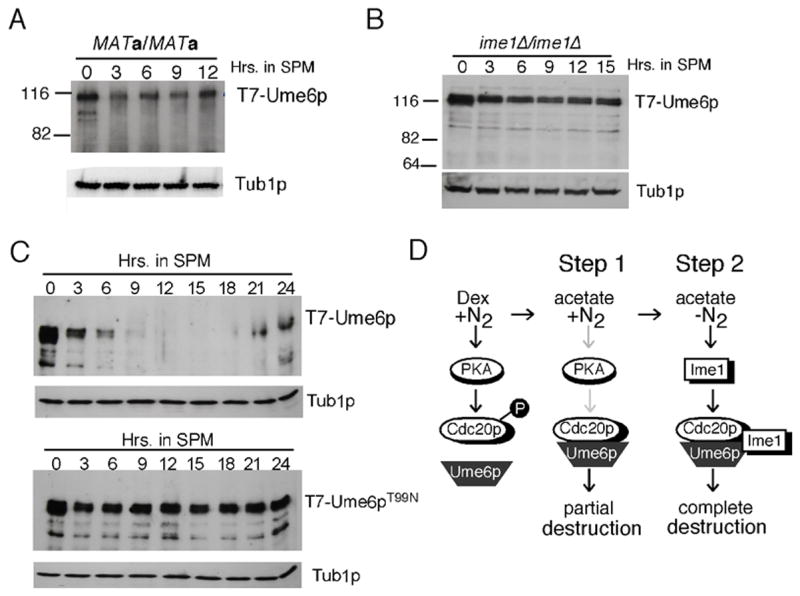
Ume6p destruction requires meiotic induction and Ime1p association. Panel A. T7-Ume6p levels were monitored by Western blot analysis in a MATa/MATa diploid (RSY303) following transfer to SPM (in hrs). Panel B. T7-Ume6p levels were monitored by Western blot analysis in an ime1Δ diploid (RSY1128) as just described in Panel A. Panel C. T7-Ume6p levels were monitored by Western blot analysis in wild type (RSY1079) and UME6T99N (RSY1395) diploid strains as described in Panel A. Panel D. Model for two-step destruction of Ume6p. The presence of dextrose and nitrogen (left panel) stimulates PKA which in turn phosphorylates Cdc20p. This modification prevents Cdc20p-Ume6p interaction. Growth on a non-fermentable carbon source (acetate) and high nitrogen (+N2) reduces PKA activity (grey arrows) reducing Cdc20p phosphorylation leading to partial degradation of Ume6p (Step 1). Step 2 (right panel) requires meiotic entry and Ime1p induction. A proposed ternary complex between Ime1p, Ume6p and Cdc20p completes Ume6p degradation.
Using a two-hybrid type experiment, a previous study found that Ime1p and Ume6p interacted and that this interaction was inhibited by a single amino acid substitution in Ume6p (T99N) (Bowdish et al., 1995). To determine if a direct interaction with Ime1p was required for Ume6p destruction, the T99N mutation was introduced into T7-UME6. This mutant was able to repress the spo13-URA3 fusion gene indicating it was functional (data not shown). Western blot analysis revealed that Ume6pT99N was not destroyed following meiotic induction (Fig. 7C). The analysis of the Ume6pT99N culture revealed that cells remained mononucleated throughout the timecourse and no spore wall assembly was observed (data not shown). Finally, Ume6p destruction was independent of Rim11p and Rim15p (Fig. S1), two protein kinases that phosphorylate Ume6p (Malathi et al., 1997; Xiao and Mitchell, 2000) and/or Ime1p (Rubin-Bejerano et al., 2004). These results indicate that Ime1p interaction is required for the meiosis-specific destruction of Ume6p.
Discussion
Similar to differentiation pathways in higher eukaryotes, the successful completion of meiosis and spore morphogenesis requires several waves of transient transcription. This study found that the induction of the early meiotic gene (EMG) class requires the destruction of Ume6p transcriptional repressor. Ume6p destruction requires Cdc20p, an activator of the ubiquitin ligase termed the anaphase promoting complex/cyclosome (APC/C). Ume6p destruction occurs in two steps. Approximately 50% reduction in Ume6p levels is observed in cells switched from a fermentable to a non-fermentable carbon source. This system is controlled by Ras and cAMP-dependent protein kinase A (PKA) through inhibitory phosphorylation of Cdc20p. The second step completely eliminates Ume6p and requires meiotic induction and direct association with Ime1p.
A role for APC/CCdc20 in controlling meiotic gene transcription is consistent with other examples of non-cell cycle related activities for the APC/C. In Drosophila, several APC/C subunits localize to post-mitotic neuromuscular synapses and are required to restrain their growth (van Roessel et al., 2004). In addition, the oncoprotein SnoN, a negative regulator of the SMAD pathway, is destroyed in an APC/C dependent manner following TGFß stimulation (Stroschein et al., 2001). Given that the HDAC-Sin3p-DNA binding protein complex functions in other development contexts (Murai et al., 2004; Wotton et al., 2001), the inactivation of HDAC activity through Ume6p destruction may represent a common mechanism by which repression is removed during cellular differentiation.
Second destruction system
Although stabilized early in meiosis, the destruction box mutant (Ume6pdb1Δ) is still destroyed 18 hrs following shift to sporulation medium (Fig. 4B). This finding suggests two possibilities. First, APC/CCdc20 may select one of the additional D-box motifs (Fig. 4A) as a default strategy to ensure meiotic induction. Alternatively, another destruction system may be in place to serve as a backup to APC/CCdc20. Mutating D-box2 and D-box3, in addition to D-box1 did not affect Ume6p destruction compared to the single mutant alone (data not shown). Mutating the non-canonical D-box4 destabilized Ume6p in vegetative cells and therefore could not be tested in this assay. In addition, the APC/C activator Hct1p/Chd1p does not mediate this second system (data not shown). However, this putative second system appears to at least require Cdc20p function as it is not activated in the cdc20-1 mutant strain at restrictive temperature (see Fig. 2A). Therefore, we can not rule out an additional role for Cdc20p in both the early and late degradation systems.
Ume6p destruction is required for meiotic gene induction
Several studies have suggested that meiotic induction requires that Ume6p is converted from a repressor to an activator by association with the meiotic inducer Ime1p (Bowdish et al., 1995; Pnueli et al., 2004; Rubin-Bejerano et al., 1996; Steber and Esposito, 1995). Ume6p destruction could be incorporated into this model if it was used as a mechanism to curtail transcriptional induction. However, several findings argue against this possibility. First, early meiotic mRNAs are relatively unstable (T1/2 ~5–10 min, (Surosky and Esposito, 1992) making it less unlikely that a delay in peak EMG mRNA accumulation would occur hours after the destruction of the trans-activator complex. Second, inactivating Cdc20p resulted in a coincident return of Ume6p and reestablishment of EMG repression. Finally, the delay of EMG induction observed with Ume6pdb1Δ is also inconsistent with the Ime1-Ume6 activator model. In addition, this result also argues against a low, undetectable pool of Ume6p remaining at EMG promoters. Rather, these results support an alternative mechanism that Ume6p destruction is required for meiotic gene induction.
Ume6p destruction; a two-step process
Step 1
This study found that Ume6p levels are reduced in cultures utilizing a non-fermentable carbon source. In addition, maintenance of Ume6p levels in dextrose grown cultures requires the Ras pathway. How does PKA protect Ume6p from destruction? Following DNA damage, PKA phosphorylates Cdc20p on two residues, S52 and S88, resulting in cell cycle arrest due to the failure to destroy the anaphase inhibitor Pds1p (Searle et al., 2004). Interestingly, serine 88 appears to provide most of the regulatory activity in the DNA damage response. Conversely, we find that phosphorylation of S52, not S88, is important for protecting Ume6p from destruction in dextrose medium. Given our finding that Ume6p interacts with Cdc20p in acetate, but not dextrose medium, suggests that PKA phosphorylation inhibits Cdc20p activity at the level of substrate association. These results are similar to those obtained monitoring Cdc20p-Clb2p interaction following DNA damage (Searle et al., 2004). Therefore, our findings are consistent with a model that high PKA activity in dextrose cultures prevents Ume6p-APC/CCdc20 interaction thus protecting Ume6p from destruction (Fig. 7D). Reduced PKA activity associated with acetate cultures (Portela et al., 2003) permits this interaction allowing the partial degradation of Ume6p. Why is there a partial reduction in Ume6p in non-dextrose cultures? Normally, yeast cultures will switch to ethanol as a carbon source one dextrose is completed. Utilizing a non-fermentable carbon source places the cell in a more physiologically competent state to enter meiosis. As Ume6p destruction is required for EMG induction and meiotic progression, its partial down regulation upon the switch to non-fermentable carbon may permit more rapid elimination of the factor thereby increasing the efficiency of meiotic induction.
Step 2
The second step, which completely depletes the cell of Ume6p, occurs upon meiotic entry. An obvious candidate for the meiotic signal that triggers Ume6p destruction was Ime1p. Ime1p is necessary for the initiation of the earliest meiotic events (Kassir et al., 1988) and interacts with Ume6p using two-hybrid and in vitro pull down studies (Bowdish et al., 1995; Malathi et al., 1997; Rubin-Bejerano et al., 1996). Our study demonstrates that Ime1p is required for Ume6p destruction. Moreover, a single amino acid substitution in Ume6p that prevents Ime1p association prevents meiotic destruction. Therefore, a combination of low PKA activity and Ime1p association insures that Ume6p destruction only occurs in the correct cell type (diploid) and low PKA activity associated with medium lacking nitrogen and a fermentable carbon source. It is unclear how Ime1p affects Ume6p accessibility and/or Cdc20p function to allow final Ume6p destruction.
Histone acetylation and the histone transacetylase Gcn5p are required for meiotic gene induction (Burgess et al., 1999; Pnueli et al., 2004). The final destruction of Ume6p would disrupt the Sin3p-Ume1p-Rpd3p HDAC complex thus permitting Gcn5p-dependent acetylation of EMG promoters. It is interesting to note that Ume6p does not appear to be required for re-establishing EMG repression as SPO13 mRNA levels are reduced prior to the recovery of Ume6p (Fig. 1C). Similarly, Ume1p and the cyclin C-Cdk8p kinase (a.k.a. Srb11-Srb10) are not required to re-establish repression (Cooper and Strich, 2002; Mallory and Strich, 2003). These findings suggest that another system is installed to repress EMG transcription as the cell completes meiosis and forms quiescent spores. Similar to differentiated tissues in higher systems, yeast spores can remain quiescent for extended periods without losing viability. Therefore, this system installed late in meiosis may be less responsive, but more sturdy, than Ume6p-dependent repression.
Experimental Procedures
Strains, plasmids and media
Detailed strain genotypes are listed in Table S1. All oligonucleotides used for site directed mutagenesis are listed in Table S2. The temperature-sensitive cdc20-1 strain RSY809 was made by back crossing H20c1a5 into our background eight times. RSY1056 (CDC16-TAP) was constructed by introducing the TAP cassette into the carboxyl terminal of CDC16 into protease deficient strain BJ5459. The ten amino acid T7 epitope tag was introduced 12 amino acids internal to the UME6 initiator ATG in plasmid p5905 (CEN LEU2) (Strich et al., 1994) by sight directed mutagenesis (Kunkel, 1985) to make pMM285. The destruction box mutations replaced the conserved arginine and leucine (RxxL) with alanines (AxxA). All mutant derivatives of UME6 were introduced into the chromosome using the pop in, pop out strategy (Rothstein, 1991) to replace the wild-type allele. All strain constructions were verified by PCR. Gene disruptions were accomplished as described (Longtine et al., 1998) and verified by PCR. The spo13-URA3 reporter gene plasmid pMS49 was described previously (Strich et al., 1994). The CLB1-3HA epitope tagged plasmid pKC426 (CEN TRP1) was constructed by PCR amplification of a chromosomally tagged CLB1 allele (Cooper et al., 2000). Medium containing the analog 5-fluoro-orotic acid (5-FOA) was prepared as described (Boeke et al., 1984). Growth and sporulation conditions were accomplished as previously described (Cooper et al., 1997).
Meiotic and Mitotic Timecourse Experiments
Meiotic timecourse experiments were conducted essentially as described (Cooper et al., 1997) at the temperature indicated in the text. The hydroxyurea (HU) arrest/release was conducted at a concentration of 200 mM at 23° for two hrs. as described (Guacci et al., 1997). Cell cycle progression was monitored by scoring for separated nuclei (anaphase) following HU release using 4’,6-diamidino-2-phenylindole (DAPI) staining. Quantitation of meiosis I and II was achieved by analyzing DAPI stained cells for the presence of two and four nuclei, respectively. At least 200 cells were counted per timepoint.
Western/Northern blot analysis
Protein extracts were prepared using a glass beat cell lysis protocol followed by an S100 clarifying centrifugation step as described (Arcangioli and Lescure, 1985). Western blot analyses of T7-tagged Ume6p and derivatives were conducted with 50μg total soluble protein. Due to the basic nature of Ume6p, the transfer step was conducted for 90 min. at 100V in 25mM CAPS (pH 10.0), 20% methanol. The alternative protein extraction protocol to visualize T7-Ume6p entailed boiling a cell pellet derived from four ml sporulation culture (5 x 107 cells/ml) in 40μl 2X sample buffer (Maniatis et al., 1982). Co-immunoprecipitation studies were accomplished using one mg of soluble protein as described (Cooper et al., 2000). Total mRNA was prepared as previously described (Strich et al., 1989) for Northern blot analysis. Probes were generated using the Prime-It II kit (Strategen) as described by the manufacturer. Quantitation of Northern blot signals was accomplished using phosphor-imaging (FUJI Inc). Western blot signals were detected using goat anti-mouse secondary antibodies conjugated to alkaline phosphatase (Sigma) and the CDP-Star chemiluminescence kit (Tropix, Bedford, MA). Quantitation of Western blot signals was performed with a Image Station 4000R (Kodak Inc.) using Molecular Imaging Software (4.0.5). For all comparative Northern and Western blot analyses, the membranes were treated with the same probe at the same time and the resulting signals developed to the same extent. Statistical analysis of Ume6p and Ume6pdb1Δ levels in cultures harboring various Cdc20p derivatives was accomplished using the unpaired t test.
In vitro ubiquitylation assays
The APC/C complex was purified from yeast extracts derived from a protease deficient strain (BJ5459) utilizing tandem affinity purification (TAP) tagged Cdc16p, a core component of this ubiquitin ligase as described previously (Passmore et al., 2003). The ligase was incubated with E. coli produced ubiquitin conjugating enzyme 6His-Ubc4p and in vitro transcription/translation produced Cdc20p (Promega Inc.). The Ume6p and Ume6pdb1Δ substrates were synthesized by in vitro transcription/translation but in the presence of 35S-methionine. The ubiquitylation reactions were conducted for the times indicated with fixed amount of Cdc20p (2.5 μl) or with increasing amounts of APC/C activator (2.5 and 5 μl) for 90 min. The reactions were stopped by addition of 2X sample buffer and separated by SDS PAGE. The gels were fixed, soaked in Amplify® (Amersham Biosciences), then dried and subjected to autoradiography.
Supplementary Material
Acknowledgments
We thank D. Barford, J. Broach, V Guacci, E.A. Jones, Y. Sanchez, M. Solomon and W. Zachariae for strains and plasmids. We also thank M. Solomon and J. Burton for helpful discussions concerning the in vitro ubiquitylation assays. This research was supported by the National Institutes of Health grant CA57842 to R.S. and the American Cancer Society (CCB106162) to K.F.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arcangioli B, Lescure B. Identification of proteins involved in the regulation of yeast iso-1-cytochrome C expression by oxygen. EMBO. 1985;4:2627–2633. doi: 10.1002/j.1460-2075.1985.tb03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5"-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bowdish KS, Mitchell AP. Bipartite structure of an early meiotic upstream activation sequence from Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2172–2181. doi: 10.1128/mcb.13.4.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdish KS, Yuan HE, Mitchell AP. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol Cell Biol. 1994;14:7909–7919. doi: 10.1128/mcb.14.12.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdish KS, Yuan HE, Mitchell AP. Positive control of yeast meiotic genes by the negative regulator UME6. Mol Cell Biol. 1995;15:2955–2961. doi: 10.1128/mcb.15.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, Zoller M, Powere S, Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987;48:789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Ajimura M, Kleckner N. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc Natl Acad Sci U S A. 1999;96:6835–6840. doi: 10.1073/pnas.96.12.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev. 2001;15:2381–2395. doi: 10.1101/gad.917901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol Cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Coluccio A, Bogengruber E, Conrad MN, Dresser ME, Briza P, Neiman AM. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:1464–1475. doi: 10.1128/EC.3.6.1464-1475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Egeland DE, Mallory MJ, Jarnik M, Strich R. Ama1p is a Meiosis-Specific Regulator of the Anaphase Promoting Complex/Cyclosome in yeast. Proc Natl Acad Sci USA. 2000;97:14548–14553. doi: 10.1073/pnas.250351297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Smith JS, Strich R. Stress and developmental regulation of the yeast C-type cyclin UME3 (SRB11/SSN8) EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Strich R. Saccharomyces cerevisiae C-type cyclin UME3/SRB11 is required for efficient induction and execution of meiotic development. Euk Cell. 2002;1:67–76. doi: 10.1128/EC.01.1.66-74.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson IA, Roth S, Artavanis-Tsakonas S. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae. J Cell Biol. 1995;129:725–737. doi: 10.1083/jcb.129.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M, Nadjar-Boger E, Capone R, Sagee S, Hashimshoni T, Kassir Y. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol Gen Genet. 1996;253:278–288. doi: 10.1007/s004380050323. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it's not just for mitosis any more. Genes & Development. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y, Granot D, Simchen G. IME1, a positive regulator of meiosis in S. cerevisiae. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M, Byers B, Esposito RE, Mitchell AP. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1997. pp. 889–1036. [Google Scholar]
- Longtine MS, McKenzie Ar, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Malathi K, Xiao Y, Mitchell AP. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory MJ, Strich R. Ume1p represses meiotic gene transcription in S. cerevisiae through interaction with the histone deacetylase Rpd3p. J Biol Chem. 2003;278:44727–44734. doi: 10.1074/jbc.M308632200. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Mitchell AP, Driscoll SE, Smith HE. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2104–2110. doi: 10.1128/mcb.10.5.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai K, Naruse Y, Shaul Y, Agata Y, Mori N. Direct interaction of NRSF with TBP: chromatin reorganization and core promoter repression for neuron-specific gene transcription. Nucleic Acids Res. 2004;32:3180–3189. doi: 10.1093/nar/gkh550. Print 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HD, Luche RM, Cooper TG. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 1992;20:1909–1915. doi: 10.1093/nar/20.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, McCormack EA, Au SW, Paul A, Willison KR, Harper JW, Barford D. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. Embo J. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Edry I, Cohen M, Kassir Y. Glucose and nitrogen regulate the switch from histone deacetylation to acetylation for expression of early meiosis-specific genes in budding yeast. Mol Cell Biol. 2004;24:5197–5208. doi: 10.1128/MCB.24.12.5197-5208.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela P, Van Dijck P, Thevelein JM, Moreno S. Activation state of protein kinase A as measured in permeabilised Saccharomyces cerevisiae cells correlates with PKA-controlled phenotypes in vivo. FEMS Yeast Res. 2003;3:119–126. doi: 10.1111/j.1567-1364.2003.tb00147.x. [DOI] [PubMed] [Google Scholar]
- Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. In: Abelson JN, Simon MI, editors. Methods in Enzymology. San Diego, CA: Academic Press Inc; 1991. pp. 281–301. [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional represssor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Sagee S, Friedman O, Pnueli L, Kassir Y. The in vivo activity of Ime1, the key transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae, is inhibited by the cyclic AMP/protein kinase A signal pathway through the glycogen synthase kinase 3-beta homolog Rim11. Mol Cell Biol. 2004;24:6967–6979. doi: 10.1128/MCB.24.16.6967-6979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah SM, Nasmyth K. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma. 2000;109:27–34. doi: 10.1007/s004120050409. [DOI] [PubMed] [Google Scholar]
- Schwab M, Neutzner M, Mocker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. Embo J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Schulze Lutum A, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin protolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Searle JS, Schollaert KL, Wilkins BJ, Sanchez Y. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat Cell Biol. 2004;6:138–145. doi: 10.1038/ncb1092. [DOI] [PubMed] [Google Scholar]
- Sia RA, Mitchell AP. Stimulation of later functions of the yeast meiotic protein kinase Ime2p by the IDS2 gene product. Mol Cell Biol. 1995;15:5279–5287. doi: 10.1128/mcb.15.10.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Simchen G. Are mitotic functions required in meiosis? Genetics. 1974;76:745. doi: 10.1093/genetics/76.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber CM, Esposito RE. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc Natl Acad Sci U S A. 1995;92:12490–12494. doi: 10.1073/pnas.92.26.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R, Slater MR, Esposito RE. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R, Surosky RT, Steber C, Dubois E, Messenguy F, Esposito RE. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- Stroschein SL, Bonni S, Wrana JL, Luo K. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 2001;15:2822–2836. doi: 10.1101/gad.912901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surosky RT, Esposito RE. Early meiotic transcripts are highly unstable in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:3948–3958. doi: 10.1128/mcb.12.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell. 2004;119:707–718. doi: 10.1016/j.cell.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Washburn BK, Esposito RE. Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol Cell Biol. 2001;21:2057–2069. doi: 10.1128/MCB.21.6.2057-2069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Knoepfler PS, Laherty CD, Eisenman RN, Massague J. The Smad transcriptional corepressor TGIF recruits mSin3. Cell Growth Differ. 2001;12:457–463. [PubMed] [Google Scholar]
- Xiao Y, Mitchell AP. Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol Cell Biol. 2000;20:5447–5453. doi: 10.1128/mcb.20.15.5447-5453.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


