Abstract
Although the integrase (IntDOT) of the Bacteroides conjugative transposon CTnDOT has been classified as a member of the tyrosine recombinase family, the reaction it catalyzes appears to differ in some features from reactions catalyzed by other tyrosine recombinases. We tested the ability of IntDOT to cleave and ligate activated attDOT substrates in the presence of mismatches. Unlike other tyrosine recombinases, the results revealed that IntDOT is able to perform ligation reactions even when all the bases within the crossover region are mispaired. We also show that there is a strong bias in the order of strand exchanges during integrative recombination. The top strands are exchanged first in reactions that appear to require 2 bp of homology between the partner sites adjacent to the sites of cleavage. The bottom strands are exchanged next in reactions that do not require homology between the partner sites. This mode of coordination of strand exchanges is unique among tyrosine recombinases.
INTRODUCTION
Bacteroides spp. are Gram-negative colonic bacteria that harbor a number of mobile genetic elements (1,2). Mobile elements called conjugative transposons (CTns) play a significant role in the transfer of antibiotic resistance genes (3,4). Integration of the Bacteroides conjugative transposon CTnDOT into the chromosome requires the joined ends of the conjugative transposon (attDOT), an attB sequence in the chromosome, a Bacteroides host factor and the CTnDOT integrase protein (IntDOT) (4,5). Recently, we showed that IntDOT belongs to the tyrosine family of recombinases (6), but the mechanism of integration and excision of CTnDOT appears to be different from that proposed for phage λ and other tyrosine recombinases (6–8).
Tyrosine recombinases utilize a topoisomerase I-like mechanism to mediate DNA rearrangements between two pairs of DNA strands (9–14). The recombination reaction can be divided into two steps and involves two staggered single-strand exchanges between DNA partners on either side of a short DNA sequence called the core or crossover region. In the first step, the active site tyrosine cleaves one strand of each DNA partner at one end of the crossover region. The products of this step are a 3′-phosphotyrosyl linkage of the enzyme to the DNA and a free 5′-hydroxyl group (15,16). The 5′-hydroxyl group from one DNA strand mediates a nucleophilic attack on the phosphotyrosyl bond of its partner. The two strands are exchanged between the DNA partners forming a Holliday Junction (HJ) (15,17,18). This intermediate is resolved in the second step of the reaction into recombinants by cleavage and strand exchanges between DNA partners at the other end of the crossover region (15). Homology between DNA partners within the crossover region is an obligate condition for the recombination reaction. The generally accepted view has been that the homology requirement in λ Int-catalyzed recombination is linked to the strand-swapping and ligation steps of the reaction (9,19–22).
The HJ is a central intermediate in recombination reactions mediated by tyrosine recombinases. However, this intermediate is extremely short-lived and usually does not accumulate during the reaction. Therefore, the kinetics of its appearance and disappearance have been difficult to study. In any recombination reaction that proceeds via a HJ, there are two possible reaction pathways: the HJ can be formed by cleavage and exchange of the top DNA strands first and then resolved by the second cleavage and exchange of the bottom DNA strand. Alternatively, the bottom strands can undergo recombination first, to generate a HJ which is later resolved by the top DNA strand exchange. Previous work has established that some tyrosine recombinase-mediated reactions, like FLP recombination (23–26), do not have a defined order of strand exchange. On the other hand, the λ Int and Xer systems show strong preferences for cleavage and exchange of one strand first during recombination (15,27–29).
DNA sequence analyses of attDOT and several attB sites yielded a consensus sequence TTTGCNNNNN (5). The TTTGC motif is completely conserved suggesting it is important for recombination. However, the 5 bp non-homologous sequence varies from site to site and recombination can occur between partner sites where all 5 bp are different. Initially the borders of the 5 bp non-homologous sequence were assumed to define the putative staggered sites of cleavage. However, using cleavage assays, we recently showed that one of the cleavage sites is actually 2 bp to the left, between the T and G, in the region of homology (6). The fact that one pair of cleavage sites lies within a region conserved in all att sites could have implications for the mechanism of cleavage and ligation of the top strands.
In this article, we show that the IntDOT recombination reaction proceeds through a HJ intermediate. We also show that there is a strong bias in the order of strand exchanges during the integrative recombination reaction. The top DNA strands are exchanged first. Using cleavage and ligation assays we show that IntDOT is able to cleave and ligate activated attDOT substrates in the presence of mismatches within the crossover region. We also report that the 2 bp region of homology between attB and an attDOT sequence on the left side of the crossover region is required for efficient recombination. Finally, we show that recombination occurs by sequential homology-dependent and homology-independent steps.
MATERIALS AND METHODS
Plasmids and oligonucleotides
The plasmids and oligonucleotides used in this work are described in Supplementary Data, Tables 2 and 3.
Site-directed mutagenesis of pUC19attDOT
The mutations were made in the 2 bp region of homology that is conserved in all known CTnDOT att sites. Mutations were made using the Stratagene Quickchange Mutagenesis Kit as described by the supplier. Primers carrying the specific mutations are shown in Table 3, Supplementary Data. The attDOT regions of the plasmid were sequenced to confirm the presence of the desired mutation and to insure that there were no additional mutations.
In vitro recombination reaction
One of the attB DNA strands was 5′ end-labeled with 32P and purified using G-25 Amersham Biosciences columns. The attB DNA substrate was prepared by mixing the labeled strand and the unlabeled complementary strand, at a 1:5 molar ratio and annealing them in an annealing buffer (0.1 M KCl, 10 mM Tris–HCl pH 8) by heating to 90°C and cooling to 25°C at a rate of 2°C/min. The attB DNA substrates containing a nick in either the bottom or top DNA strand were prepared by mixing a labeled strand and two unlabeled oligonucleotides that were complementary to the labeled one, at ratios of 1:5:5, respectively and annealing them as described above. All suicide attB substrates were phosphorylated at the 5′ end at the cleavage position to prevent rejoining after strand exchange. The attDOT and attB substrates were then incubated in a 20 μl volume containing 30 mM Tris–HCl (pH 7.4), 2 mM DTT, BSA (70 μg/ml), 2.6% glycerol and 50 mM KCl. Proteins and DNA were added to the final concentrations: IntDOT, 70 nM; IHF, 40 nM; attB DNA, 2 nM; and attDOT plasmid DNA, 3 nM. Samples were incubated for 2 h at 37°C. All samples were loaded onto 1% agarose gels and subjected to electrophoresis. The gels were exposed to phosphorimager screens and the reaction efficiencies were quantified using a Fuji FLA-3000 phosphorimager and Fujifilm Image Gauge software (Macintosh v.3.4). Results were calculated where indicated by dividing a total amount of label present in the product band by the total amount of label present in the substrate and product bands, multiplied by 100 to give percent recombination.
Denaturing agarose gel analysis
The intact attB substrates and attB substrates that contained a single-stranded DNA nick at the IntDOT cleavage site in the bottom strand were radiolabeled at the 5′ end. The recombination reactions were performed with attDOT DNA substrates using IntDOT as described above. Samples were electrophoresed through a 1.5% agarose gel in 50 mM NaOH and 1 mM EDTA, at 35 V for 16 h. The gel was then neutralized by soaking in 500 ml of 1 mM HCl for 1 h and dried on a vacuum slab drier and exposed to phosphorimager screens.
The 2D gel analysis is described in Supplementary Data.
Restriction digest analysis of the products of IntDOT recombination reaction
The attB DNA substrates, labeled as indicated in the text, were recombined with attDOT in a double volume of the standard recombination reaction. The reaction was terminated by addition of 200 μl of phenol/chloroform/isoamyl alcohol mixture (25:24:1). The mixture was vortexed and spun in a microcentrifuge and the aqueous phase was transferred to a fresh tube. The DNA was ethanol-precipitated and re-suspended in a 50 μl 1× reaction buffer and digested with SspI endonuclease. Samples were electrophoresed through a 1% agarose gel at 90 V for 100 min. The gel was dried on a vacuum slab drier and exposed to phosphorimager screens.
Detection of covalent DNA/protein complexes
A double size standard recombination reaction was terminated by the addition of 0.1 vol of 10% SDS, followed by addition of 0.5 ml of A1 buffer (10 mM Tris–HCl pH 7.5, BSA 20 μg/ml, calf thymus DNA 20 μg/ml, 1% SDS). The sample was vortexed and incubated at 37°C for 4 min. Fifty microliters of 2.5 M KCl was added to the mixture which was placed on ice for 10 min. The precipitate was collected by centrifugation for 2 min at 4°C and re-suspended in 1 ml of cold B buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 100 mM KCl). The supernatant fraction was saved and the pellet was washed twice. The final precipitate was re-suspended in 1 ml of buffer containing 10 mM Tris–HCl pH 7.5, 1 mM EDTA, 100 mM NaCl, 10 mM MgCl2 and tRNA (20 μg/ml). DNA was precipitated by addition of 2.5 vol of 100% ethanol. The pellet was washed in 70% ethanol, air-dried, and re-suspended in 20 μl of water. The pellet and the supernatant were electrophoresed through a 1% agarose gel.
Cleavage and ligation assays
The cleavage and ligation assays have been described previously (6) and are included in Supplementary Data.
RESULTS
A bias for top strand exchange is observed with an attB suicide substrate
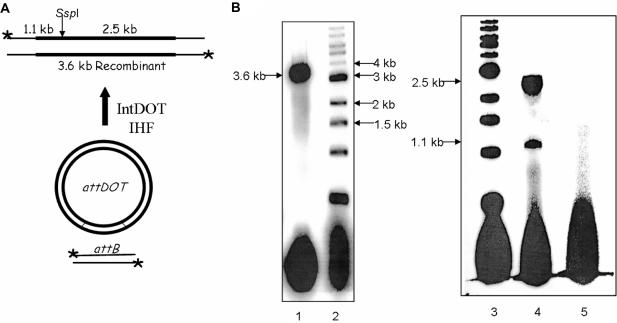
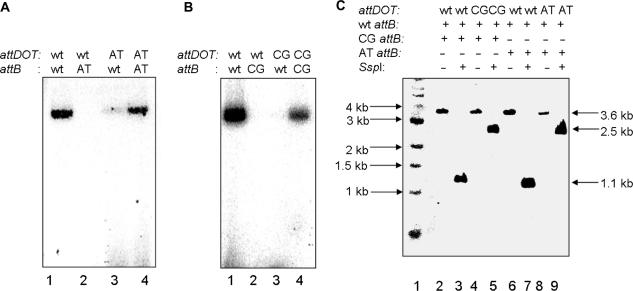
We developed an in vitro recombination assay based on the one previously described for λ Int (15). The IntDOT in vitro recombination reaction occurs between a circular attDOT site on a supercoiled plasmid and a linear attB DNA. The product of the reaction is a linear DNA fragment containing both attL and attR sites (Figure 1A). IntDOT was able to catalyze an efficient recombination reaction in vitro. Up to 45% of the labeled attB substrate underwent recombination. As expected, the linear product of the reaction migrated as a 3.6 kb linear fragment on an agarose gel. The recombinant should contain a unique SspI site. As shown in Figure 1B, digestion of the recombinant with SspI resulted in the formation of two linear DNA fragments of the expected lengths (1.1 and 2.5 kb).
Figure 1.
Restriction digest analysis of IntDOT recombinant. (A) The recombination reaction mediated by IntDOT occurs in the presence of E. coli IHF. The reaction occurs between a linear radiolabeled (asterisk) attB DNA and an attDOT site cloned on a plasmid. The product of the reaction is a linear 3.6 kb long recombinant. Cleavage by SspI should generate fragments of 1.1 and 2.5 kb. (B) Agarose gel analysis of recombination products. Lane 1, 3.6 kb long linear recombinant. Lane 2, 1 kb ladder. Lane 3, 1 kb ladder. Lane 4, products of the recombination reaction digested with SspI enzyme. Lane 5, the recombination reaction with no IntDOT and IHF added.
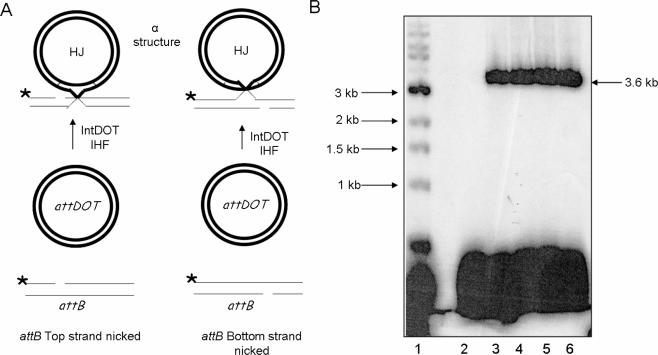
Recombinant product should be formed by two sets of strand exchanges between attDOT and attB sites. The top DNA strands can be cleaved and exchanged first to form a HJ, followed by a second cleavage, exchange and ligation of the bottom DNA strands. Alternatively the bottom strands can undergo strand exchange first, to generate a HJ which is resolved by the top DNA strand exchanges. To determine whether CTnDOT recombination proceeds by ordered strand exchanges, we used attB suicide substrates that contained a nick in one DNA strand at one of the sites of IntDOT cleavage (15). Introduction of the nick at a cleavage site should block recombination because the enzyme cannot form the phosphotyrosyl–protein intermediate. On the other hand, an intact attB strand can be cleaved and exchanged with its DNA partner. Inhibition of the second strand exchange by a nick should lead to the formation of a HJ with a single-strand crossover. Since one DNA partner is circular and the other is linear, a HJ should appear as an ‘α structure’ and would be expected to migrate in an agarose gel like a relaxed plasmid (Figure 2A) (15,28). If the nick is introduced in the top attB strand, an α intermediate should be observed if the bottom strand is exchanged first. Alternatively, if a nick is introduced in a bottom strand, an α intermediate should be observed only if the top strand is exchanged first. If either top or bottom strand can be exchanged first, an α intermediate should be observed with both attB nicked substrates (Figure 2A).
Figure 2.
Recombination reactions with attB suicide substrates nicked on either the top or bottom DNA strand. (A) Recombination reactions with attB nicked suicide substrates. The attB substrates contain a nick at the site of cleavage in the top (left) or bottom (right) strands. Strand exchange of an attB substrate with a nick in the top strand occurs if the bottom strands are exchanged first (left). If the nick is in the bottom strand of attB, a strand exchange occurs if the top strand is exchanged first (right). The strand with a nick placed in a cleavage site should not undergo recombination. The HJ intermediate has a form of an α structure. (B) Agarose gel analysis of recombination reactions with attB substrates that contain a ssDNA nick at the IntDOT cleavage site in the top or in the bottom DNA strands. Lane: 1, 1 kb ladder. Lanes 2 and 3 are reactions with a nick in the top (lane 2) or bottom (lane 3) attB strand. Lane 4 is a reaction with an intact attB. Lanes 5 and 6 are attB substrates that contained a nick in the top (lane 5) or bottom (lane 6) strand that were treated with T4 DNA ligase to seal the nick.
We were unable to observe the product of recombination in an in vitro recombination reaction between supercoiled attDOT and labeled attB containing a nick in the top DNA strand (Figure 2B, lane 2). To show that lack of a product in the reaction is due to the presence of a nick and not because the substrates were not fully hybridized, we performed reactions with the same suicide attB substrates that were previously incubated with T4 DNA ligase and ATP to seal the nick. The ligated substrate was able to form the product (Figure 2B, lanes 5 and 6). In the recombination reaction with an attB substrate having a nick in the bottom DNA strand, we observed the formation of a product (Figure 2B, lane 3) with the same electrophoretic mobility as the linear recombinant (Figure 2B, lanes 3 and 4). The results of this experiment indicate that there is a bias in strand exchange during the recombination reaction and suggest that the top strand is exchanged first. In the recombination reaction with a suicide attB substrate, we expected to trap a HJ as an α structure intermediate. However, it was surprising to find that actual intermediate had the same electrophoretic mobility as the linear recombinant while an α structure would have migrated slower in an agarose gel.
Analysis of the predicted CTnDOT HJ
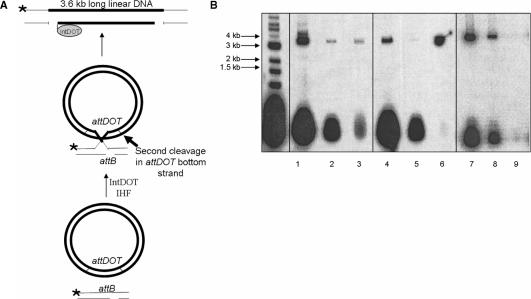
Digestion of an α structure with a restriction enzyme containing a single restriction site should form a nicked HJ (15,28), with expected migration mobility close to the linear DNA of the same size in agarose gel. Surprisingly, digestion of the product generated two DNA fragments 1.1 and 2.5 kb long which were the same size as SspI generated fragments from a complete recombination reaction (Figure 3, lanes 1 and 3). This result was unexpected and suggested that after formation of a HJ by a first round of strand exchanges a second DNA cleavage occurs on the bottom strand of attDOT DNA that is not followed by DNA ligation. The structure of this product is shown in Figure 4A.
Figure 3.
Restriction digest analysis of the product of the IntDOT-mediated recombination reaction with suicide attB. Recombination reaction with an intact attB substrate labeled at both ends (lane 2) and digested with SspI (lane 1). Reactions with bottom strand nicked attB (lane 4) and digested with SspI (lane 3). Molecular weight standards are in lane 5.
Figure 4.
Isolation of covalent IntDOT/DNA complexes, using the SDS/KCl precipitation method. (A) Recombination reaction with an attB substrate nicked in the bottom DNA strand. After formation of the HJ, a second DNA cleavage occurs in the attDOT site in the bottom DNA strand that is not followed by DNA ligation. The product of this reaction is a linear 3.6 kb long DNA fragment containing two nicks in the bottom DNA strand and protein attached to the 3′ attDOT DNA end. (B) Recombination reaction with intact attB substrates (lanes 1–3), with attB containing a nick in IntDOT cleavage site in the bottom DNA strands (lanes 4–6) and with attB containing a nick in IntDOT cleavage site on the bottom DNA strand where proteinase K was added before SDS/KCl precipitation (lanes 7–9). Lanes 1, 4 and 7 contain untreated reactions. Lanes 2, 5 and 8 contain supernatants after SDS/KCl precipitation. Lanes 3, 6 and 9 contain pellets after SDS/KCl precipitation.
The partially blocked IntDOT recombination reaction forms a covalent DNA/protein complex
If IntDOT is able to perform a second cleavage on the bottom attDOT strand during the reaction with suicide attB substrate, the protein should remain covalently attached to the 3′-DNA end (Figure 4A). Treatment of the reaction mixture with SDS/KCl should precipitate only proteins or DNA that is covalently bound to a protein (30,31). Recombination reactions with suicide and intact attB substrates were performed and the reactions were divided in half. In one sample, proteins were precipitated from the reaction mixtures and pelleted by centrifugation. The supernatant fractions were saved. The untreated sample and supernatant and pellet from the SDS/KCl-treated sample were then analyzed on an agarose gel. (Figure 4B) The results of the experiment revealed that stable covalent DNA/protein complexes are formed during recombination reaction with suicide attB substrates. When intact attB substrates were used DNA precipitation was negligible (Figure 4B, lanes 1, 2 and 3). We were able to precipitate DNA from the reaction mixture only when attB suicide substrates were used (Figure 4B, lanes 4, 5 and 6). Proteinase K treatment was used to show that this precipitation was due to the covalent linkage of the protein to DNA (Figure 4B, lanes 7, 8 and 9). Our results indicate that after formation of a HJ in the reaction with a suicide attB substrate, IntDOT is able to perform a second DNA cleavage in the bottom attDOT DNA strand that is not followed by DNA ligation.
The top DNA strand is exchanged during IntDOT-mediated recombination with attB suicide substrates
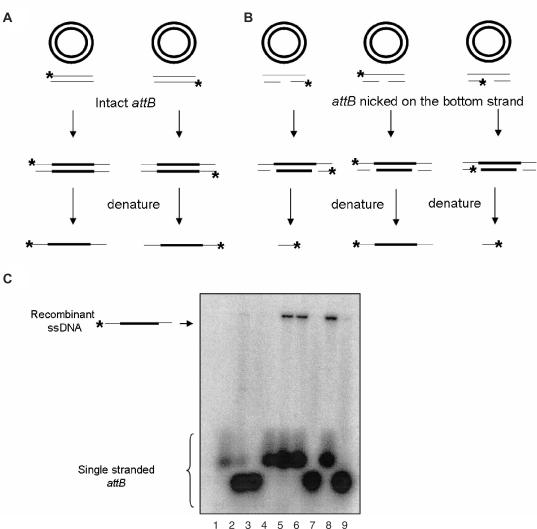
In order to determine if the product of the reaction with attB containing a nick in the bottom strand is a DNA molecule with a single-stranded crossover on the top strand, we used denaturing agarose gel analysis. We used intact attB substrates and attB substrates with a preexisting nick in the bottom DNA strand, where only one strand was radiolabeled. The recombination reactions were performed and each DNA strand was then separated on an agarose gel under denaturing conditions (Figure 5C). As predicted, denaturation of the product of the reaction with intact attB yields a long single-stranded DNA, when either the bottom or the top strand was labeled (Figure 5C, lanes 5 and 6). Denaturation of the product of the recombination reaction with an attB suicide substrate, in which the label was placed on the top strand, yielded long single-stranded DNA fragments of the same size as recombinant DNA (Figure 5C, lane 8). When the bottom attB strands were labeled, we were unable to detect any product (Figure 5C, lanes 7 and 9) indicating that the bottom DNA strand did not undergo strand exchange.
Figure 5.
Denaturing agarose gel analysis of products.(A) Predicted products of recombination reactions with intact attB. (B) Predicted products of recombination reactions with suicide attB. (C) Denaturing agarose gel analysis of intact attB reaction with either attB strand labeled yields intact a 3.6 kb single-stranded DNA. The product of a strand exchange with a suicide attB substrate with the top strand labeled should yield a 3.6 kb single-stranded DNA. If either of the bottom strands is labeled, a 40-base single-stranded DNA should be generated. Denatured substrates: lane 1, denatured top strand of intact attB; lane 2, denatured left, bottom strand of attB; lane 3, denatured right, bottom strand of attB; lane 4, denatured bottom strand of intact attB. Recombination reactions: lane 5, with intact attB with the top strand labeled; lane 6, with intact attB with the bottom strand labeled, lane 7, with suicide attB with the bottom right strand labeled; lane 8, with suicide attB with the top strand labeled; lane 9, with suicide attB with the bottom left strand labeled.
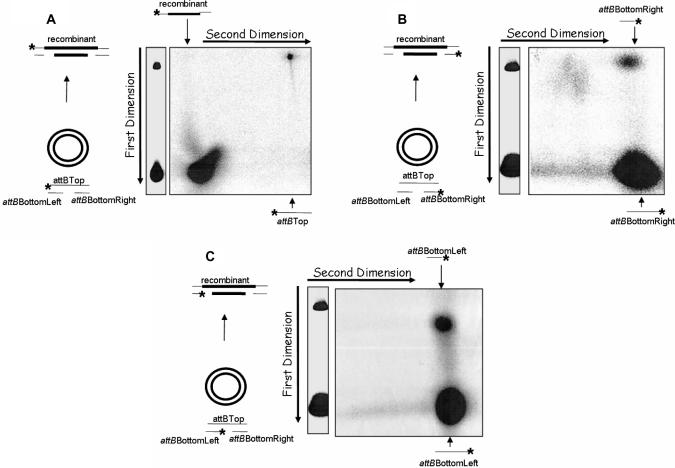
To confirm that the product of IntDOT-mediated recombination reaction with a suicide attB is a DNA molecule with the top strand exchanged, we used 2D gel analysis. The recombination reactions were performed with bottom strand nicked attB substrates containing a label either on the top or on one of the bottom strands. After separation on a native agarose gel, the single strands of each fragment were separated by electrophoresis in the second dimension under denaturing conditions (Figure 6A–C). If the top strand was labeled and if only the top DNA strand underwent recombination we would expect a long, ssDNA that migrated slowly in the second dimension on a denaturing agarose gel. By contrast, if either bottom strand was labeled we would expect short DNA fragments that migrated faster in the agarose gel, because the bottom DNA strand did not undergo recombination. Our analysis showed that denaturation of the product of these reactions released a long top strand (Figure 6A) and short bottom attB strands (Figure 6B and C). These results indicated that only the top strand was exchanged, whereas the bottom strand did not undergo strand exchange.
Figure 6.
Two-dimensional gel analysis of the products generated by IntDOT-mediated recombination. The attB substrates contain a nick in the IntDOT cleavage site in the bottom DNA strand. attB DNA was radiolabeled at one 5′ end (asterisk), incubated with attDOT DNA in a standard recombination reaction and treated as described in Materials and Methods section. (A) The top attB DNA strand labeled. (B) The right bottom strand (attBBottomRight oligo) labeled. (C) The left bottom attB strand (attBBottomLeft oligo) labeled.
Introduction of heterology at the left side of the crossover region blocks CTnDOT recombination reaction
Previously we demonstrated that the IntDOT top strand cleavage site lies in a region of homology between the T and G of the conserved TTTGCNNNNN sequence. Thus, both the cleavage and ligation reactions involving the top strand take place in an environment where base pairing can occur. In order to determine whether the homology at the left side of the CTnDOT crossover region is required for the recombination reaction to occur or, alternatively, if the reaction can occur if the sites have heterology on the left side, we performed recombination reactions using intact attB and attDOT substrates that contained a GC to CG or AT mutation in the region of homology. The reactions were performed with equimolar concentrations of labeled attB substrates. The efficiency of recombination reaction was calculated as described in Materials and Methods section.
As shown in Table 1, when mutated attB sites (attBCG or attBAT) recombined with wild-type attDOT, integration was 1000 times less efficient than for the reactions with wild-type attB. A similar effect was observed for the reactions where mutated attDOTCG recombined with wild-type attB (Table 1, Figure 7B). IntDOT was able to catalyze the recombination reaction between wild-type attB site and the attDOT site containing the AT mutation, but the efficiency of this reaction was severely depressed (Table 1). To exclude the possibility that the decrease in the recombination activity between wild-type and mutated att site could be due to the lack of Int binding to the mutated sequence, not to the disruption of the homology between attB and attDOT, we tested recombination efficiency between attDOT and attB containing the same mutations in the crossover region [Figure 7A (lane 4) and B (lane 4)]. As shown in Table 1, the recombination frequency between attDOT and attB containing the same mutation was similar to the wild-type recombination activity.
Table 1.
Effect of core mutations on IntDOT recombination
| attDOT sites | attB sites | ||
|---|---|---|---|
| Wild-type attB | attBCG | attBAT | |
| Wild-type attDOT | 33.00% ± 12% | 0.02% ± 0.04% | 0.04% ± 0.03% |
| attDOTCG | 0.10% ± 0.5% | 13.50% ± 8% | – |
| attDOTAT | 2.00% ± 1% | – | 26.50% ± 10% |
The recombination frequencies were calculated as described in Materials and Methods section. All experiments were repeated three times.
Figure 7.
IntDOT recombination reactions with mutated attB and attDOT substrates. Mutations were introduced in the 2 bp region of homology in the left side of IntDOT crossover region. The GC sequence was changed to AT or CG, respectively. (A) Recombination between sites with GC to AT mutations. Lane 1, wild-type attDOT and wild-type attB; lane 2, wild-type attDOT and attBAT; lane 3, wild-type attB and attDOTAT; lane 4, attDOTAT and attBAT. (B) Recombination between sites with GC to CG mutations. Lane 1, wild-type attDOT and wild-type attB; lane 2, wild-type attDOT and attBCG; lane 3, wild-type attB and attDOTCG; lane 4, attDOTCG and attBCG. (C) A competition assay. Equimolar concentrations of wild-type attB labeled on the top strand and mutated attB labeled on the bottom strand were added to the reaction with wild-type attDOT or mutated attDOT, respectively. Reactions were digested with SspI enzyme. Digestion of the recombinant with a top strand labeled should give 1.1 kb DNA fragment. Digestion of the recombinant with the bottom strand labeled should give 2.5 kb fragment.
We also performed competition assays in which an equimolar mixture of duplex wild-type attB labeled on the top strand and duplex mutated attB (attBAT or attBCG) labeled on the bottom strand were incubated with wild-type attDOT in the reaction mixture. Reaction products were digested with SspI and analyzed on an agarose gel. Digestion of the recombinant with the top DNA strand labeled should give only a 1.1 kb fragment that could be detected on the gel, while digestion of the recombinant containing the label on the bottom DNA strand should generate 2.5 kb detectable fragment only. The only labeled band was the 1.1 kb long DNA fragment (Figure 7C, lanes 3 and 7). This indicated that only wild-type attB site was able to recombine with a wild-type attDOT site. Alternatively, when we incubated the wild-type attB labeled on the top strand and mutated attBCG labeled on the bottom strand with mutated attDOTCG, we were able to detect only the 2.5 kb long DNA fragment (Figure 7C, lane 5). Similar results were obtained when GC sequence was changed to AT (Figure 7C, lane 9). These results indicated that attB substrates could only recombine with attDOT carrying the same 2 bp sequence on the left side of the crossover region. From our data we conclude that 2 bp homology on the left side of the crossover region is critical for the IntDOT recombination reaction while the remaining 5 bp are not conserved.
IntDOT efficiently cleaves mispaired attDOT phosphorothioate DNA substrates
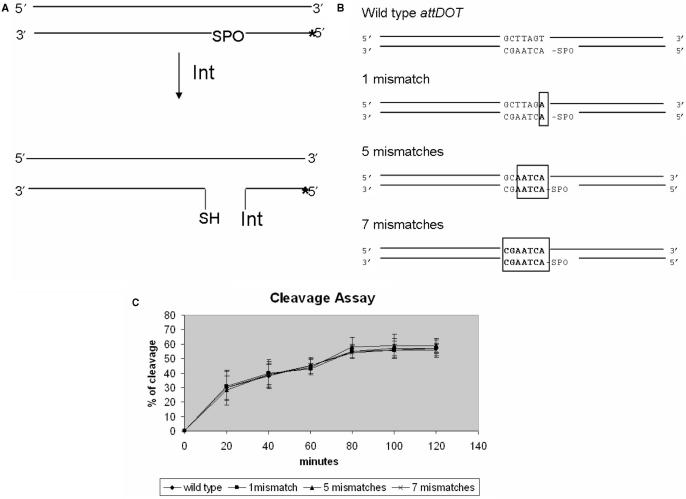
The in vitro cleavage and ligation assays allowed us to determine whether lack of base paring within the IntDOT crossover region affected those steps in the recombination reaction. To determine the cleavage rates of the wild-type attDOT DNA substrate and attDOT substrates containing mismatches introduced in the IntDOT crossover region, a phosphorothioate cleavage assay was used (6). In this assay, the oxygen which forms a phosphotyrosyl bond with the catalytic tyrosine is replaced by sulfur. The cleavage reaction leaves behind a 5′ SH group. This reaction is essentially irreversible and the protein remains covalently attached to the 3′ end of the DNA (Figure 8A). This product can be detected as a shift in migration distance of attDOT DNA due to attachment of the protein. We used attDOT cleavage substrates containing 1, 5 and 7 mismatches introduced in the CTnDOT crossover region (Figure 8B). The yield of all those reactions was comparable with wild-type reaction efficiency (Figure 8C). Results of the cleavage assay showed that the CTnDOT cleavage reactions can occur in the presence of mismatches.
Figure 8.
Phosphorothioate cleavage assay. (A) The labeled attDOT DNA substrate (asterisk) contains a 5′-bridging phosphorothioate linkage at the site of cleavage. If the enzyme cleaves the DNA, it becomes covalently bound to the substrate DNA. This reaction is irreversible. (B) The attDOT substrates used in cleavage assays. (C) Results of the phosphorothioate cleavage assay with wild-type attDOT sequence and attDOT sequence containing 1, 5 and 7 missmatches within the crossover region. The reaction efficiencies were calculated as described in Materials and Methods section.
IntDOT ligates mispaired attDOT ligation substrates
Studies of FLP recombination target sites (FRT) demonstrated that homology in the crossover region is required to successfully complete the ligation step of recombination reaction (22). It was also shown that the efficiency of the ligation reaction of λ Int was sensitive to mismatches during DNA hairpin formation and very sensitive to the homology at the site of ligation during resolution of HJ (32). Since it was previously shown that IntDOT can perform the recombination reaction when all five bases on the right side of the crossover region are different between two att sites (5), we expected that IntDOT could perform the ligation reaction without homology between DNA partners. We tested if IntDOT is sensitive to mismatches in an in vitro p-nitrophenol (pNP) assay (6,33,34). The ligation substrate, a pNP oligonucleotide, mimics the activated 3′-phosphotyrosine intermediate that is formed after substrate cleavage. The enzyme binds the DNA, forms a phosphodiester bond, and releases the pNP (Figure 9A). We measured the ligation rates of the wild-type attDOT DNA substrate and the attDOT substrates containing 2, 5 and 7 mismatches introduced in the IntDOT crossover region (Figure 9B). The ligation product can be detected by formation of a 44-base DNA product on a denaturing gel (33,34). In the experiments reported here, the amount of product was measured as a function of time. The results of these experiments are shown in Figure 9C. IntDOT was able to perform ligation on the pNP attDOT substrates containing up to 7 mismatches. This reaction was less efficient than the wild-type reaction, although half of the substrate was ligated, which was ∼70% of the ligation efficiency of strands that were complementary to the partner site (Figure 9C). We repeated the same experiment for λ Int and we found that this enzyme was not able to perform the ligation reaction on a similar activated attP substrate containing a single mismatch within the crossover DNA sequence (Figure 9C). The ability of λ Int to ligate attP pre-activated substrate was reestablished after the homology was restored (data not shown). Our observation that the IntDOT-mediated ligation reaction can occur in the absence of base pairing, while other tyrosine recombinases require complete base pairing for ligation, indicates that the architecture of IntDOT catalytic site has significant differences from the other well-studied tyrosine recombinases.
Figure 9.
Ligation assay. (A) The labeled DNA substrate (asterisk) contains a p-nitrophenol linkage incorporated at the 3′ site of cleavage, which mimics phosphotyrosyl bond between IntDOT and DNA. This substrate can be recognized and ligated by λ or CTnDOT integrase. (B) attDOT and attP substrates used in ligation assays. (C) Results of ligation assays with variations of attDOT (left) and λ attP (right). The reaction efficiencies were calculated as described in Materials and Methods section.
DISCUSSION
A molecular analysis of the mechanism of the IntDOT-mediated recombination reaction depends on the ability to block the reaction at intermediate steps. In tyrosine recombinase reactions, HJs are intermediates but do not usually accumulate because they are rapidly resolved to recombinants or substrates (35,36). We also did not observe a detectable amount of HJ formed during in vitro recombination (Figure 1B). To determine if the CTnDOT reaction proceeds via a HJ intermediate, we needed to find a way to block the reaction at the intermediate step. Several modifications of reaction conditions that stop tyrosine recombinase reaction at the HJ step have been described. These include incorporation of a non-bridging phosphorothioate in the DNA at the site of cleavage (28) or use of small synthetic peptides that are known to trap HJs (37). We were unable to detect HJs with these methods.
We found that the introduction of a nick in either the top or bottom attB strand influenced IntDOT recombination (15). A nick at the top strand inhibited the reaction without accumulation of an intermediate, while a nick at the bottom strand promoted the accumulation of an intermediate (Figure 2). However, we demonstrated that the product is not a free HJ. After the first cleavage and strand exchange in the reaction with the nicked top strand, IntDOT performs a second cleavage on the bottom attDOT DNA strand which is not followed by ligation. The product of this reaction was a linear DNA molecule that contained two nicks in the bottom strand and the protein covalently attached to one of the 3′ DNA ends (Figure 4). Other well-studied tyrosine recombinases do not form a similar product (15), indicating that IntDOT recombination differs from other well-studied tyrosine recombinases reactions. It is known that some tyrosine recombinase-mediated reactions, like FLP recombination (23–26), do not have a defined order of strand exchanges. On the other hand, λ Int and Xer systems show strong preferences for the initial cleavage and exchange of one particular pair of strands (15,27–29). We interpret our results to be strong evidence that CTnDOT integrative recombination proceeds by sequential DNA exchanges in which the top DNA strands in each site are cleaved and exchanged first.
Most tyrosine recombinases display a strong requirement for sequence identity in the crossover region between DNA partners for completion of the recombination reaction (10,19,38). For example Bauer et al. (19) showed that the single base-pair mismatch adjacent to the cleavage site in the crossover region in the top strand of a λ att site strongly inhibited the reaction. When both sites contained the homologous mutated base pair, recombination was restored. Kitts and Nash (39) demonstrated that heterology introduced on the right side of the crossover region provided a partial block of the reaction and lead to the accumulation of HJs. They explained this need for homology by a branch-migration model, which involves migration of the junction branch point between the sites of Int cleavage. After the strand exchange, the lack of base pairing between DNA partners provides an energetic barrier to the branch migration process and blocks the second exchange. Landy and coworkers (20,21,32) proposed an alternative strand-swapping-isomerization model, in which homology is sensed during the formation of the new bases between DNA partners, after cleavage and prior to ligation. They showed that the first-strand exchange in λ recombination depends on complementary base pairing between DNA partners. They also demonstrated that λ Int ligation is affected by the presence of mismatches at the site of ligation. Studies with FLP and Cre recombinases showed similar requirements for homology (22,40–43)
In some cases, cleavage and ligation reactions have been shown to occur in regions of heterology. The topological analysis of the recombination products from in vitro assays with supercoiled plasmids containing two heterologous FRT sites on the same molecule revealed that the Flp-mediated reaction can occur in the presence of heterology but the products are rapidly resolved back to substrates (44). Sherratt and coworkers (45) used an in vivo Xer-mediated recombination system, which used two plasmids that contained mutated recombination target sites (cer). They found that formation of the HJ product occurred with some substrates that contained mismatched bases. Experiments monitoring recombination of synthetic HJs that contained varying junction positions showed that ligation occurs between sites with mispaired bases. Ligation was most efficient when the mispaired base was away from the cleavage point and least efficient when it was adjacent to the cleavage site (45,46). However, the conditions used for these experiments did not employ substrates or intermediates that are normal participants in the respective recombination reactions.
Our previous analyses showed that CTnDOT att sites contain a 2 bp region of homology on the left side and a 5 bp non-homologous region on the right side of the crossover sequence. This finding suggests that recombination mediated by IntDOT is different from reactions catalyzed by other well-studied tyrosine recombinases and proceeds by homology-dependent and homology-independent strand exchanges. We propose that the IntDOT recombination reaction proceeds by the following general steps. First an intasome composed of IntDOT, IHF (or the Bacteroides host factor) and attDOT DNA is formed. Four monomers of IntDOT bind to the four sites in attDOT and attB that flank the crossover sites. The complex captures and undergoes synapsis with a partner attB site. This step could be similar to synapsis by other tyrosine recombinases, and would not require homology in the crossover regions of attDOT and attB (31,47). During the first cleavage, strand exchange and ligation reaction, only two out of four monomers are active. Results described in this article strongly indicate that the first cleavage and ligation reactions occur in the conserved region of homology in the top DNA strands (Figures 5 and 6). This event appears to require homology at the left side of the crossover region because reactions with attB and attDOT substrates that contained GC to CG or AT mutations in the 2 bp region of homology are severely depressed (Table 1). It is possible that the homology dependence at this step occurs by the strand-swapping-isomerization model according to which homology is sensed during the formation of the new bases between DNA partners before the first ligation and formation of a HJ (20). Alternatively, the homology could be required only for ligation of the exchanged strands after cleavage.
The next step of the reaction would involve an isomerization of the HJ, where the complex changes conformation that activates the other two partner IntDOT monomers at the sites of the second strand exchange. Since the remaining five bases on the right side of the crossover region are not complementary, the isomerization step would be homology independent because movement of the DNA occurs in the absence of complementary base pairs. It is possible that formation of the two complementary base pairs on the left side is important for the proper isomerization of the junction. The second pairs of cleavage, strand exchange and ligation reactions would form the recombinant products. This step is homology independent as shown in this work and requires ligation of exchanged strands under conditions where they cannot form Watson–Crick base pairs with the new partner. The final step of the IntDOT-mediated recombination would be dissociation of the complex with the recombinant sites containing a five base region of heteroduplex DNA between the sites of strand exchanges.
We demonstrated here that IntDOT cleavage and ligation reactions can occur in the presence of mismatches (Figures 8 and 9). Although ligation and cleavage assays were not performed in the context of recombination complexes we confirmed that IntDOT efficiently cleaves mispaired attDOT phosphorothioate substrates (Figure 8). In the in vitro ligation assay, IntDOT was able to ligate DNA substrates in the presence of seven mismatches in the crossover region. In contrast, introduction of a single mismatch in a pNP attP substrate completely abolished λ ligation in a similar experiment (Figure 9C). These findings emphasize drastically different properties of the two enzymes.
IntDOT was classified as a member of the tyrosine recombinase family, although our previous results indicated that the catalytic core of the protein seems to have somewhat different organization than other well-studied tyrosine recombinases (6). In this article, we demonstrate that the coordination by the strand exchange catalyzed by IntDOT appears to be distinct from ones used by other tyrosine recombinases such as λ Int. Our observations indicate that the first strand exchange in IntDOT-mediated recombination is a homology-dependent step, while second round of recombination occurs in the region of heterology where neither cleavage, exchange or ligation of the bottom DNA strand require homology. To our knowledge, IntDOT is the only tyrosine recombinase to use this type of two-step mechanism. It will be interesting to determine whether this feature is found in other recombinases encoded by other conjugative transposons.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr S. Silverman and Dr C. Hoebartner for helping us with synthesis of a phosphorothioate substrate for the cleavage assay and Dr A. Burgin for providing p-nitrophenol substrate for ligation assays. We also thank Dr J. DiChiara for providing IntDOT protein, Dr D. Schlesinger for providing pUC19attDOT and L. Rajeev for her helpful input on IntDOT recombination mechanism analysis. This work was supported by U.S. National Institute of Health grant GM 28717. Funding to pay the Open Access publication charges for this article was provided by NIH Grant GM 28717.
Conflict of interest statement. None declared.
REFERENCES
- 1.Salyers AA, Shipman JA. 11: Getting in touch with your prokaryotic self: mammal-microbe interactions. In: Staley JT, Reysenbach A.-L, editors. Biodiversity of Microbial Life. Wiley-Liss, Inc.; 2002. pp. 315–341. [Google Scholar]
- 2.Macy JM, Probst I. The biology of gastrointestinal bacteroides. Annu. Rev. Microbiol. 1979;33:561–594. doi: 10.1146/annurev.mi.33.100179.003021. [DOI] [PubMed] [Google Scholar]
- 3.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive resistance gene transfer among Bacteroides spp and between Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 2001;67:561–568. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whittle G, Shoemaker NB, Salyers AA. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 2002;59:2044–2054. doi: 10.1007/s000180200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng Q, Paszkiet BJ, Shoemaker NB, Gardner JF, Salyers AA. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 2000;182:4035–4043. doi: 10.1128/jb.182.14.4035-4043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malanowska K, Salyers AA, Gardner JF. Characterization of a conjugative transposon integrase, IntDOT. Mol. Microbiol. 2006;60:1228–1240. doi: 10.1111/j.1365-2958.2006.05164.x. [DOI] [PubMed] [Google Scholar]
- 7.Whittle G, Salyers AA. Bacterial transposons-an increasingly diverse group of elements. In: Streips UN, Yasbin RE, editors. Modern Microbial Genetics. 2nd. New York: Wiley-Liss, Inc; 2002. pp. 387–427. [Google Scholar]
- 8.Scott JR, Churchward GG. Conjugative transposition. Annu. Rev. Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 9.Bauer CE, Gardner JF, Gumport RI, Weisberg RA. The effect of attachment site mutations on strand exchange in bacteriophage lambda site-specific recombination. Genetics. 1989;122:727–736. doi: 10.1093/genetics/122.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Duyne GD. A structural view of tyrosine recombinases site-specific recombination. In: Craig NL, Craige R, Gellert M, Lambowitz, editors. Mobile DNA II. Washington, D.C.: ASM press; 2002. pp. 93–117. [Google Scholar]
- 11.Bushman W, Thompson JF, Vargas L, Landy A. Control of directionality in lambda site specific recombination. Science. 1985;230:906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng C, Kussie P, Pavletich N, Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 13.Craig NL, Nash HA. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983;35:795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- 14.Duckett DR, Murchie AI, Diekmann S, von Kitzing E, Kemper B, Lilley DM. The structure of the Holliday junction, and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 15.Nunes-Duby SE, Matsumoto L, Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987;50:779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- 16.Pargellis CA, Nunes-Duby SE, de Vargas LM, Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J. Biol. Chem. 1988;263:7678–7685. [PubMed] [Google Scholar]
- 17.Hsu PL, Ross W, Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980;285:85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holliday R. The induction of mitotic recombination by mitomycin c in ustilago and Saccharomyces. Genetics. 1964;50:323–335. doi: 10.1093/genetics/50.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer CE, Gardner JF, Gumport RI. Extent of sequence homology required for bacteriophage lambda site-specific recombination. J. Mol. Biol. 1985;181:187–197. doi: 10.1016/0022-2836(85)90084-1. [DOI] [PubMed] [Google Scholar]
- 20.Nunes-Duby SE, Azaro MA, Landy A. Swapping DNA strands and sensing homology without branch migration in lambda site specific recombination. Curr. Biol. 1995;5:139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- 21.Nunes-Duby SE, Yu D, Landy A. Sensing homology at the strand-swapping step in lambda excisive recombination. J. Mol. Biol. 1997;272:493–508. doi: 10.1006/jmbi.1997.1260. [DOI] [PubMed] [Google Scholar]
- 22.Zhu XD, Pan G, Luetke K, Sadowski PD. Homology requirements for ligation and strand exchange by the FLP recombinase. J. Biol. Chem. 1995;270:11646–11653. doi: 10.1074/jbc.270.19.11646. [DOI] [PubMed] [Google Scholar]
- 23.Luetke KH, Sadowski PD. Determinants of the position of a Flp-induced DNA bend. Nucleic Acids Res. 1998;26:1401–1407. doi: 10.1093/nar/26.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Tonozuka T, Jayaram M. Mechanism of active site exclusion in a site-specific recombinase: role of the DNA substrate in conferring half-of-the-sites activity. Genes Dev. 1997;11:3061–3071. doi: 10.1101/gad.11.22.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Tribble G, Jayaram M. Resolution of tethered antiparallel and parallel holliday junctions by the Flp site-specific recombinase. J. Mol. Biol. 2000;296:403–419. doi: 10.1006/jmbi.1999.3472. [DOI] [PubMed] [Google Scholar]
- 26.Luetke KH, Sadowski PD. The role of DNA bending in Flp-mediated site-specific recombination. J. Mol. Biol. 1995;251:493–506. doi: 10.1006/jmbi.1995.0451. [DOI] [PubMed] [Google Scholar]
- 27.Cassell GD, Segall AM. Mechanism of inhibition of site-specific recombination by the Holliday junction-trapping peptide WKHYNY: insights into phage lambda integrase-mediated strand exchange. J. Mol. Biol. 2003;327:413–429. doi: 10.1016/s0022-2836(03)00058-5. [DOI] [PubMed] [Google Scholar]
- 28.Kitts PA, Nash HA. Bacteriophage lambda site-specific recombination proceeds with a defined order of strand exchanges. J. Mol. Biol. 1988;204:95–107. doi: 10.1016/0022-2836(88)90602-x. [DOI] [PubMed] [Google Scholar]
- 29.Arciszewska LK, Sherratt DJ. Xer site-specific recombination in vitro. EMBO J. 1995;14:2112–2120. doi: 10.1002/j.1460-2075.1995.tb07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trask DK, DiDonato JA, Muller MT. Rapid detection and isolation of covalent DNA/protein complexes: application to topoisomerase I and II. EMBO J. 1984;3:671–676. doi: 10.1002/j.1460-2075.1984.tb01865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nunes-Duby SE, Matsumoto L, Landy A. Half-att site substrates reveal the homology independence and minimal protein requirements for productive synapsis in lambda excisive recombination. Cell. 1989;59:197–206. doi: 10.1016/0092-8674(89)90881-7. [DOI] [PubMed] [Google Scholar]
- 32.Lee SY, Landy A. The efficiency of mispaired ligations by lambda integrase is extremely sensitive to context. J. Mol. Biol. 2004;342:1647–1658. doi: 10.1016/j.jmb.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Kazmierczak RA, Swalla BM, Burgin AB, Gumport RI, Gardner JF. Regulation of site-specific recombination by the C-terminus of {lambda} integrase. Nucleic Acids Res. 2002;30:5193–5204. doi: 10.1093/nar/gkf652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodfield G, Cheng C, Shuman S, Burgin AB. Vaccinia topoisomerase and Cre recombinase catalyze direct ligation of activated DNA substrates containing a 3′-para-nitrophenyl phosphate ester. Nucleic Acids Res. 2000;28:3323–3331. doi: 10.1093/nar/28.17.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Echols H, Green L. Some properties of site-specific and general recombination inferred from int-initiated exchanges by bacteriophage lambda. Genetics. 1979;93:297–307. doi: 10.1093/genetics/93.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enquist LW, Nash H, Weisberg RA. Strand exchange in site-specific recombination. Proc. Natl Acad. Sci. USA. 1979;76:1363–1367. doi: 10.1073/pnas.76.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klemm M, Cheng C, Cassell G, Shuman S, Segall AM. Peptide inhibitors of DNA cleavage by tyrosine recombinases and topoisomerases. J. Mol. Biol. 2000;299:1203–1216. doi: 10.1006/jmbi.2000.3829. [DOI] [PubMed] [Google Scholar]
- 38.Azaro MA, Landy A. The isomeric preference of Holliday junctions influences resolution bias by lambda integrase. EMBO J. 1997;16:3744–3755. doi: 10.1093/emboj/16.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitts PA, Nash HA. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987;329:346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- 40.Guo F, Gopaul DN, Van Duyne GD. Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc. Natl Acad. Sci. USA. 1999;96:7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J, Jayaram M. Role of partner homology in DNA recombination. Complementary base pairing orients the 5′-hydroxyl for strand joining during Flp site-specific recombination. J. Biol. Chem. 1995;270:4042–4052. doi: 10.1074/jbc.270.8.4042. [DOI] [PubMed] [Google Scholar]
- 43.Dixon JE, Sadowski PD. Resolution of synthetic chi structures by the FLP site-specific recombinase. J. Mol. Biol. 1993;234:522–533. doi: 10.1006/jmbi.1993.1608. [DOI] [PubMed] [Google Scholar]
- 44.Azam N, Dixon JE, Sadowski PD. Topological analysis of the role of homology in Flp-mediated recombination. J. Biol. Chem. 1997;272:8731–8738. doi: 10.1074/jbc.272.13.8731. [DOI] [PubMed] [Google Scholar]
- 45.McCulloch R, Coggins LW, Colloms SD, Sherratt DJ. Xer-mediated site-specific recombination at cer generates Holliday junctions in vivo. EMBO J. 1994;13:1844–1855. doi: 10.1002/j.1460-2075.1994.tb06453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arciszewska L, Grainge I, Sherratt D. Effects of Holliday junction position on Xer-mediated recombination in vitro. EMBO J. 1995;14:2651–2660. doi: 10.1002/j.1460-2075.1995.tb07263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burgin A.B., Jr, Nash HA. Suicide substrates reveal properties of the homology-dependent steps during integrative recombination of bacteriophage lambda. Curr. Biol. 1995;5:1312–1321. doi: 10.1016/s0960-9822(95)00258-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.