Abstract
Human ERCC1/XPF is a structure-specific endonuclease involved in multiple DNA repair pathways. We present the solution structure of the non-catalytic ERCC1 central domain. Although this domain shows structural homology with the catalytically active XPF nuclease domain, functional investigation reveals a completely distinct function for the ERCC1 central domain by performing interactions with both XPA and single-stranded DNA. These interactions are non-competitive and can occur simultaneously through distinct interaction surfaces. Interestingly, the XPA binding by ERCC1 and the catalytic function of XPF are dependent on a structurally homologous region of the two proteins. Although these regions are strictly conserved in each protein family, amino acid composition and surface characteristics are distinct. We discuss the possibility that after XPF gene duplication, the redundant ERCC1 central domain acquired novel functions, thereby increasing the fidelity of eukaryotic DNA repair.
INTRODUCTION
During DNA replication, recombination and repair, double-stranded DNA inevitably forms three- or four-way junctions, bubbles, flaps or broken ends with single-stranded extensions. These irregular structures must be processed correctly in order to successfully complete DNA metabolism and thereby maintain genome integrity. This task is accomplished by structure-specific endonucleases specialized in pruning downstream of branch, flap or bubble structures by incision at junctions between double- and single-stranded DNA (1). Inactivation or malfunctioning of these enzymes causes genetic defects or cancer, underlying their importance in genome stability.
A remarkable class of structure-specific endonucleases in humans is XPF. The protein family is characterized by the presence of the ERCC4 domain and consists of seven members (XPF, MUS81, ERCC1, EME1, EME2, FANCM and FAAP24) (2). Only XPF and MUS81 have nuclease activity, which is mediated by the conserved core nuclease motif (ERKX3D) (3–5). Their catalytic function depends on heterodimer formation with the non-catalytic family members. XPF forms an obligate complex with ERCC1 and functions primarily in nucleotide excision repair (NER), a versatile pathway able to detect and remove a variety of DNA lesions induced by UV light and environmental carcinogens. The ERCC1/XPF heterodimer has additional roles in DNA interstrand cross-link (ICL) repair (6) and telomere maintenance (7). The symptoms of the first patient with inherited ERCC1 deficiency (8) and of a patient with a novel XPF mutation (9) are distinct from the classical NER phenotype, and underscore the pleiotropic function of ERCC1/XPF.
In contrast to eukaryotes, archaea have a single homolog of the XPF endonuclease that forms homodimers. The archaeal XPF minimally consists of the catalytic nuclease domain followed by a DNA-binding domain containing two consecutive helix-hairpin-helix motifs (HhH2 domain). Dimerization occurs between both the nuclease and HhH2 domains of each subunit (Figure 1A) (3). The strong preference of the archaeal endonucleases for 3′ flap DNA substrates is explained by the function of their individual domains. Structural data suggest a model for DNA binding where one HhH2 domain of the dimer binds to an upstream and the other to a downstream DNA duplex, sharply bending the DNA substrate and thereby allowing one active site of the dimeric nuclease domain to cleave the 3′ protruding single strand (10). Combination of mutations in nuclease and HhH2 domains that do not disrupt the dimeric interfaces but impair the individual functions (DNA cleavage or DNA binding), provide evidence for this model (11).
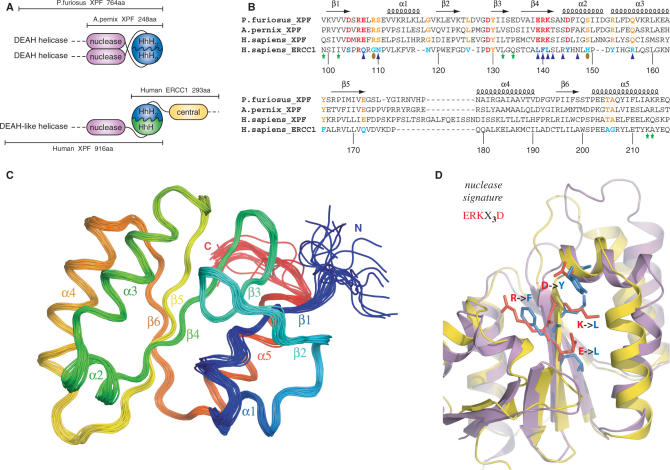
Figure 1.
(A) Domain organization of the archaeal XPF homodimeric members and the human ERCC1/XPF heterodimer. (B) Structure-based sequence alignment of the nuclease XPF domains from archaea to human and the corresponding central domain of human ERCC1. Secondary structure elements of the prototype XPF nuclease fold are indicated at the top of the sequences. Catalytic residues in the XPF nucleases are colored red and their corresponding substitutions in ERCC1 blue. Other invariant residues in XPF domains are depicted in orange, while the ERCC1 equivalents are depicted in cyan. Residues of cERCC1 perturbed largely upon XPA titration are indicated by blue triangles and those appear only in the final complex by brown ellipses. Green asterisks indicate cERCC1 residues perturbed by DNA titration. cERCC1 sequence is numbered at the bottom. (C) Ensemble of the final 20 structural conformers of cERCC1 as determined by solution NMR. Secondary structure elements and N- and C-termini are labeled. (D) Superposition of the crystal XPF nuclease structure (2bgw) from A. pernix (purple) and the solution NMR structure (2jpd) of human cERCC1 (yellow). Emphasis is given to the nuclease signature and the corresponding substitutions in cERCC1.
The DNA substrate specificity is different for the human ERCC1/XPF heterodimer. The human enzyme is known to specifically incise hairpin, bubble or splayed-arm DNA substrates (12,13), which consist of only one duplex (upstream). Previous data indicated the role of the C-terminal ERCC1 ‘canonical’ HhH2 domain in engaging the upstream DNA duplex of a hairpin substrate (14). This function requires heterodimerization with the C-terminal HhH domain of the human XPF partner to form a complex analogous to the archaeal HhH2 dimer interface (12,14). Previous structural data (12) and the data presented here illustrate the structural similarity between the human ERCC1 central domain and the archaeal XPF nuclease domain. Unlike the archaeal nuclease dimer interface, there is no evidence that the corresponding domains of human ERCC1 and XPF interact with each other in solution (Figure 1A) (12).
The catalytic function of the ERCC1/XPF endonuclease is crucial for NER. NER operates through a ‘cut and patch’ mechanism by excising and removing a short stretch of DNA containing the lesion, and subsequently restoring the genetic information by repair synthesis using the undamaged strand as the template. The incision step involves the sequential and coordinated assembly of the DNA damage sensor XPC-HR23B, the transcription/repair factor TFIIH, the architectural protein XPA, the ubiquitous ssDNA-binding protein RPA, and the two structure-specific endonucleases ERCC1/XPF and XPG, responsible for the incisions 5′ and 3′ to the damaged site, respectively (15). A main role in the progress of the reaction is attributed to XPA, which in complex with RPA probes the DNA helix conformations (16) and participates in multiple interactions with the other NER factors (15). XPA is required for the recruitment of ERCC1/XPF in the NER pre-incision complex (17,18). Consistent with this regulatory function, in vitro studies with recombinant proteins and in vivo studies using a yeast two-hybrid system have demonstrated interactions between XPA and ERCC1 (19).
Here, we report the solution structure of the ERCC1 central domain (cERCC1) and investigate its interactions with XPA and DNA at the molecular level. Using biochemical studies and NMR titration experiments, we have identified two distinct interaction surfaces of cERCC1 that mediate XPA and DNA binding. Interestingly, the two interactions can take place simultaneously to yield a cERCC1/DNA/XPA ternary complex, which in turn explains the important role of ERCC1 in targeting its catalytic XPF partner to the NER pre-incision complex.
MATERIALS AND METHODS
Cloning, protein expression and protein purification
ERCC1 central domain (cERCC1, residues 96 to 219) was PCR-amplified from a vector containing the full-length ERCC1 gene and subcloned into a pET28b (Novagen) expression vector between BamHI and XhoI sites. Downstream of the XhoI site a linker together with a his-tag has been engineered. XPA constructs were derived from a full-length XPA vector purchased from RZPD (clone ID IRAUp969B1273D6) and subcloned into a pLICHIS vector using the enzyme free cloning (EFC) strategy (20). The same procedure was followed for the GST fusion proteins, either XPA or cERCC1 (pLICHISGST vector). As a result all the EFC constructs contain an N-terminal his-tag, while the cERCC1 construct bears a C-terminal his-tag. The nucleotide sequences of the cloned DNAs were confirmed by sequencing. The cERCC1-HIS, HIS-XPA, HIS-GST-XPA and HIS-GST-cERCC1 proteins were expressed in BL21(DE3) Rosetta cells (Novagen) and were subject to two-step purification as has been described (21). When appropriate, HIS-GST-cERCC1 was cleaved with thrombin in the presence of 1 mM Ca2+, 50 mM Tris (pH 8.0) and 100 mM NaCl in order to obtain the untagged protein. Finally, the protein samples were exchanged either to 50 mM sodium phosphate (pH 5.5), 100 mM NaCl for the cERCC1 structure determination, or to 50 mM Tris (pH 8.0), 100 mM NaCl for the binding studies.
NMR spectroscopy
Multidimensional NMR experiments were carried out at 290 K on Bruker AVANCE 600 and 900 NMR spectrometers equipped with TXI triple-resonance probes in 50 mM sodium phosphate (pH 5.5), 100 mM NaCl by using the cERCC1-HIS protein. Spectra were processed using the NMRPipe software package (22) and analyzed with Sparky (23). The 1H, 15N and 13C resonance assignments are 97% complete and were made using standard triple resonance techniques, 3D NOESY-(15N, 1H)-HSQC spectra, and 3D NOESY-(13C, 1H)-HSQC spectra (both with a mixing time of 60 ms) (24). The chemical shifts have been deposited at the BMRB with accession number 15240.
Structure calculations
Automatic NOE assignment and structure calculations were performed using the program CYANA version 2.0 (25). Hydrogen bond restraints were defined when they were consistent with the secondary chemical shift data and expected NOE contacts and only for the helical parts of the protein. The set of 2669 NOE restraints determined by CYANA, together with restraints for 31 hydrogen bonds and ϕ and ψ torsion angle restraints (174) derived from TALOS (26) were used in a water refinement run according to the standard RECOORD protocol (27) utilizing CNS (28). Molecular images were generated with PyMol (29). Coordinates have been deposited at the PDB with accession code 2jpd and the structural restraints at the BMRB with entry number 15240.
EMSAs
All the DNA substrates were 5′ 32P-labeled with T4 polynucleotide kinase and purified on a polyacrylamide gel under native conditions. The nucleotide sequences for ss20, b10 and ds30 have been described before (30). Per reaction, 100 fmol of substrate was incubated with the appropriate amount of ERCC1 protein in binding buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 10% glycerol and 1 mg/ml BSA). After incubation for 30 min at 4°C, samples were loaded onto 7.5% native polyacrylamide gels containing 0.5× TBE and run at 4°C. Alternatively Electrophoretic mobility shift assay (EMSA) reactions were loaded on a 3.5% agarose gel containing 0.5% TBE, run at 4 or 20°C yielding essentially identical results. Gels were visualized and quantified using a phosphor imager (BioRad) as described before (30). For depletion experiments, 20 µl of MagneHis beads (Promega) or GST agarose beads (Sigma), were extensively washed with binding buffer, and after buffer removal the EMSA reaction was added to the beads. After 15 min incubation with regular mixing, the beads were removed from the EMSA reaction using a magnet or centrifugation and the reaction mixture was loaded on acrylamide gel or, when appropriate, the substrate was added to the depleted reaction mixture.
GST pull-down assay
GST pull-down assay was performed and quantified as described before (31) using the indicated cERCC1 concentrations and 3–5 µg of GST proteins in 150 µl of 50 mM Tris, 100 mM NaCl, 10% glycerol, 1 mM DTT, 0.2 mM PMSF and 20 mg/ml of BSA (pH 8.0). The semi-quantitative experiments were performed in 50, 150, 500, 1500 and 5000 µl buffer with a constant amount of cERCC1 and GST-XPA.
NMR titrations
cERCC1 titrations with XPA and DNA were performed on a Bruker AVANCE 700 NMR spectrometer and were monitored with 2D 1H-15N HSQC experiments. In all cases, the concentration of cERCC1 was 0.2 mM in 50 mM Tris (pH 8.0), 100 mM NaCl. XPA and DNA were dissolved in the same buffer and therefore salt and pH did not vary throughout the experiments. DNA binding was feasible only with the cERCC1 construct lacking the C-terminal his-tag, while XPA binding was identical, regardless of the his-tag presence. Normalized chemical shift changes were calculated by using the equation: δ = ([δHN]2 + [δN/6]2)0.5.
RESULTS AND DISCUSSION
Structure analysis
We have determined the solution structure of the ERCC1 central domain (cERCC1) to elucidate the molecular details of its proposed functions. We have collected a large number of distance and dihedral angle restraints that yielded an ensemble of 20 conformers with very good convergence (Figure 1C). The quality of the structure can be judged by the summary of the structural and restraint statistics given in Table 1. All the secondary structure elements are well defined including the short loops that join them. cERCC1 comprises a compact architecture and folds as a six-stranded β-sheet flanked by five α-helices on both sides. As described before (12), this fold is reminiscent of the type II restriction endonucleases, to which the catalytic domain of XPF also belongs.
Table 1.
Structural statistics of the structure ensemble of human cERCC1 (residues 96–219)
| Rmsd (Å) with respect to meana | |
| Backbone/Heavy atoms | 0.40 ± 0.04/0.89 ± 0.08 |
| Number of experimental restraints | |
| Intraresidue NOEs | 632 |
| Sequential NOEs (|i − j| = 1) | 678 |
| Medium range NOEs (1<|i − j| < 4) | 479 |
| Long-range NOEs (|i − j| > 4) | 880 |
| Total NOEs | 2669 |
| Dihedral angle restrains | 174 |
| Hydrogen bonds | 31 |
| Restraint violations | |
| NOE distances with violations >0.3 Å | 0.00 ± 0.00 |
| Dihedrals with violations >3° | 0.00 ± 0.00 |
| Rmsd for experimental restraints | |
| All distance restraints (2669) (Å) | 0.013 ± 0.005 |
| Torsion angles (174) (°) | 0.579 ± 0.077 |
| CNS energies after water refinement | |
| Evdw (kcal/mol) | −614 ± 17 |
| Eelec (kcal/mol) | −4914 ± 61 |
| Rmsd from idealized covalent geometry | |
| Bonds (Å) | 0.01 ± 0.00 |
| Angles (°) | 1.15 ± 0.03 |
| Impropers (°) | 1.34 ± 0.06 |
| Ramachandran analysis | |
| Residues in the favored regions (%) | 90.68 ± 2.04 |
| Residues in additional allowed regions (%) | 8.16 ± 1.71 |
| Residues in generously allowed regions (%) | 0.81 ± 0.65 |
| Residues in disallowed regions (%) | 0.36 ± 0.68 |
aResidues 101–215 of cERCC1.
Overall, the solution NMR structure of cERCC1 is in very good agreement with the crystal structure of the same domain (2a1i) (rmsd 1.1 Å for 108 Cα atoms). The only noticeable difference relates to the last few residues of helix α3 and the loop connecting this structure element with β5. The presence of a mercury atom (linked to C137) in the crystal structure may account for the substantial side-chain rearrangements within this loop. When compared to the crystal structures of the archaeal nucleases, helices α1, α2, and strands β1, β2 appear to be shorter in the solution structure of cERCC1. Solution cERCC1 and the crystal structures of archaeal XPF nucleases (2bgw and 1j23) contain the same number of secondary structure elements arranged in a very similar manner (rmsd 1.8 and 2.0 Å for Aeropyrum pernix and Pyrococcus furiosus nucleases, respectively, for 108 Cα atoms), although in both cases the primary sequence homology is very low (Figure 1B and D).
Remarkably, conserved residues of XPF nuclease scattered in the primary sequence superimpose structurally with residues conserved in the ERCC1 sequence family (Figures 1 and 3). Whereas the conservation in XPF is directly related to the catalytic function, ERCC1 has preserved the same fold but lacks the essential residues for catalysis. The fold similarities, coupled with the obligate nature of the heterodimerization (32), are in full agreement with the common origin of the two proteins (33).
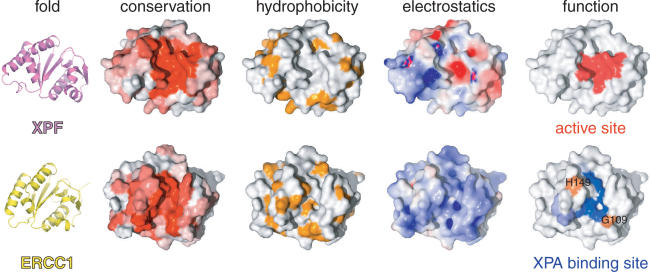
Figure 3.
Common fold, different properties and distinct functions for XPF and ERCC1. Sequence conservation either for XPF or ERCC1 proteins is colored from white (non-conserved) to red (highly conserved). The opposite face has no significant conservation for either protein. Hydrophobic side chains are colored orange. Electrostatic surface potentials were calculated using APBS (39) and colored blue for positive or red for negative charge potential. Active site residues of XPF were chosen based on previous mutational studies (5) and the XPA-binding site of ERCC1 was identified by our NMR titrations.
Minimal domains for ERCC1–XPA interaction
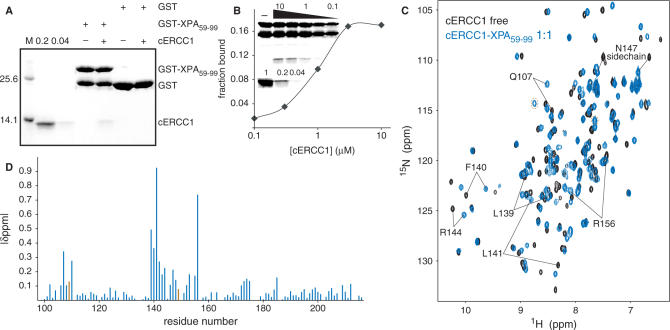
Truncation studies of ERCC1 have mapped the XPA interaction site to a region between residues 91 and 118 (34). From the cERCC1 structure and its compact fold, we predict that such truncations will have a devastating effect on the structural integrity of the central domain (96–219). For XPA, the ERCC1 interaction region seems to be located in a small stretch containing two highly conserved motifs rich in glycine and glutamic acid residues (72-GGGFILEEEEEEE-84, conserved residues are underlined) (19). We have prepared a HIS-GST-XPA construct (59–99) containing the conserved stretch, and examined its ability to interact with cERCC1 in a GST pull-down assay. Indeed, the GST XPA fusion protein was able to specifically bind cERCC1 (Figure 2A), confirming that this domain of ERCC1 is sufficient for interactions with XPA. No binding of cERCC1 to GST bound agarose beads or uncharged agarose beads was observed (Figure 2A, data not shown). This XPA peptide is unstructured (35) and appeared very sensitive to proteolysis both during overexpression in Escherichia coli and as pure protein after extensive purification (Figure 2A and B). We next performed a semi-quantitative GST pull-down assay (31) and estimated an apparent Kd of 1 µM. Since we reached only 20% binding saturation at the two highest protein concentrations (3 and 10 µM of cERCC1) under these experimental conditions, we could not determine the binding constant more accurately (Figure 2B).
Figure 2.
ERCC1–XPA interactions. (A) GST pull-down assay with 3 µg of GST or GST-XPA fusion proteins in the presence or absence of 2 µM cERCC1. Here, 0.2 and 0.04 refer to respectively 20 and 4% of the input cERCC1 protein present in the GST pull-down assay. (B) Semi quantitative GST pull-down assay showing the fraction of cERCC1 bound to 3 µg of GST-XPA at the indicated [cERCC1] (µM). The upper part of inset shows a representative GST pull-down assay, where the various cERCC1 concentrations used are depicted above (10, 3, 1, 0.3 and 0.1 µM). The lower panel shows respectively 100, 20 and 4% of cERCC1 added to the assay. (C) Chemical shift perturbation of the cERCC1 1H-15N HSQC upon complex formation with XPA. Free cERCC1 spectrum is in black and XPA-bound spectrum in blue, while brown circles in the bound spectrum indicate G109 and H149 resonances. (D) Normalized chemical shift changes between free and XPA-bound forms versus the cERCC1 sequence. The p.p.m. difference for G109 and H149 was calculated by their resonances in the free cERCC1 spectrum at pH 5.5, and are colored brown.
Functional disparity of the XPF nuclease and ERCC1 central domains
The cERCC1–XPA interaction specificity has been explored using NMR titration experiments. Titration with the HIS-XPA peptide causes extensive specific changes in the cERCC1 1H-15N HSQC NMR spectra. The titrations show slow exchange regime with respect to the NMR chemical shift timescale, in agreement with the determined affinity (Figure 2C). We have been able to assign all the amide resonances of cERCC1 in the bound form (1:1 complex stoichiometry) by using 3D NOESY-(15N, 1H)-HSQC spectra. The pattern of the backbone amide NOEs does not change substantially compared to the free cERCC1 protein. This indicates that the overall structure is maintained, and led us to conclude that the observed chemical shift perturbations arise from specific contacts with the XPA peptide.
The most significant perturbations map to the surface of the V-shaped groove of the cERCC1 structure and involve many residues that compose the corresponding DNA cleavage site of the archaeal XPF endonucleases (Figures 2D and 3). Interestingly, among the largest chemical shifts of cERCC1 upon XPA binding are those of L139, F140 and L141. These ERCC1 residues correspond to the characteristic strongly conserved ERK catalytic triad of the XPF nuclease domain (Figure 1). The rest of the significantly perturbed residues are polar or positively charged (Q107, N110, S142, R144, N147 and R156), are competent for hydrogen bonding with the XPA peptide and are more conserved than the surrounding regions of the cERCC1 V-shaped groove (Figure 3). Additionally, of special interest are the amide signals of G109 and H149, which are not observed in the free protein spectrum due to fast exchange with the solvent at pH 8 (these amide resonances are observable at pH 5.5). However, both give sharp signals at the end of the titration, an indication that the solvent exchange in the free protein is quenched in the complex. These residues reside at the rim of the V-shaped binding site and may be involved in hydrogen bonding with the XPA peptide (Figure 3).
Although ERCC1 residues forming the XPA-binding site are different from the acidic and basic residues which form the active site in XPF (5), there are no significant changes in the backbone fold and the spatial side-chain orientations in the structure remain unaffected. However, their different chemical properties result in a distinct charge and hydrophobicity distribution on the structure surface (Figure 3). While XPF displays negative charge at the DNA cleavage site (important for divalent cation coordination), the equivalent XPA-binding site in ERCC1 is neutral or slightly positively charged. Therefore, the ERCC1–XPA interaction is a combination of hydrophobic and electrostatic interactions possibly involving the glutamic acid stretch of XPA (19) and two conserved positively charged residues (R106 and R156) of cERCC1.
DNA binding by the ERCC1 central domain
Given the proposed common origin of ERCC1 and XPF (33), it can be expected that the degenerated catalytic site of ERCC1 could still bind DNA in a similar fashion as the archaeal XPF active site (12). Because our ERCC1–XPA interaction data render this unlikely, we explored whether cERCC1 is able to bind DNA as has been suggested previously (12).
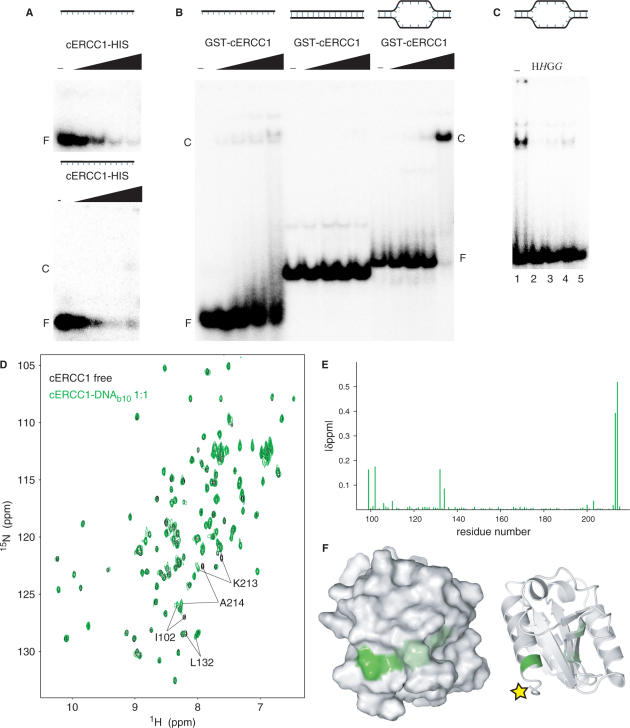
We performed an electrophoretic mobility shift assay (EMSA) where increasing amounts of cERCC1-HIS protein were added to ssDNA. While we saw a loss of free probe, we failed to see any complex formation under these conditions. If the same protein–DNA complex was loaded on a 3.5% agarose gel, we detected a weak smeary complex at the highest protein concentration (Figure 4A). However, by using a HIS-GST-cERCC1 protein, we observed formation of the complex with ssDNA (Figure 4B). This suggests that the C-terminal his-tag interferes with DNA binding. Therefore, we used the HIS-GST-cERCC1 protein, which we cleaved with thrombin to obtain the untagged protein. DNA binding with the untagged cERCC1 demonstrated a faster migrating complex with various ssDNA substrates (data not shown). HIS-GST-cERCC1/DNA complex formation is prevented by incubation of the reaction mixture with MagneHIS magnetic beads or glutathione agarose beads, which depletes HIS-GST-cERCC1 from the reaction mixture either prior to or after the addition of DNA. If beads were added after complex formation, a small but significant decrease in the amount of free probe was observed. Depletion of the HIS-GST tagged cERCC1 protein from the binding reaction confirms that the protein–DNA complex is indeed formed by the HIS-GST-cERCC1 protein (Figure 4C).
Figure 4.
DNA binding by cERCC1. (A) EMSA with a 20-mer ssDNA substrate using increasing concentrations of cERCC1-HIS protein (0, 0.04, 0.2, 1, and 5 µM), loaded on a 7.5% acrylamide gel (upper panel) or a 3.5% agarose gel (lower panel). (B) EMSA using ssDNA (20-mer), dsDNA (30 bp) and bubble10, a dsDNA with 10 unpaired bases as substrate, in the presence of 0, 0.125, 0.25, 0.5 and 1 µM HIS-GST-cERCC1. (C) Inhibition of cERCC1–b10 DNA complex formation ([cERCC1] = 0.5 µM) by depletion of HIS-GST-cERCC1 from the EMSA reaction by the addition of MagneHis beads (H) or GST agarose beads (G) prior to or after addition of DNA (italic). F: free DNA, C: cERCC1–DNA complex. (D) Chemical shift perturbation after addition of b10 DNA. The spectrum of the free protein is shown in black and the spectrum after addition of equimolar amount of b10 in green. (E) Normalized chemical shift changes between free and b10-bound forms versus the cERCC1 sequence. (F) Surface and cartoon representations of cERCC1 colored according to normalized chemical shifts. Yellow star indicates the position of the C-terminal his-tag that was interfering with DNA binding.
In agreement with earlier studies, under these conditions no binding was observed for dsDNA (12), while fairly comparable binding affinities were found for both ssDNA and bubble substrates with 10 or 20 unpaired bases. We calculated an apparent Kd of 2.5 ± 0.7 μM for the HIS-GST-cERCC1 protein bound to a bubble10 (b10) substrate in the presence of 100 mM NaCl (Figure 4B). Corroborating our results, an equilibrium binding titration experiment using fluorescence anisotropy has demonstrated ssDNA binding for the same domain with an apparent Kd of 10 μM (12). The relatively small difference in affinity can be well explained by different salt conditions or substrate used.
For the NMR titrations we performed a thrombin cleavage on HIS-GST-cERCC1, to remove the his-tag. The untagged cERCC1 1H-15N HSQC spectrum is identical to that of cERCC1-HIS, except for the two amides preceding the artificial his-tag (Supplementary Figure 1). Addition of the b10 substrate to this cERCC1 protein caused a limited number of specific perturbations that were unambiguously assigned. Again, the perturbed resonances exhibit slow exchange behavior in the NMR titrations and indicate contacts with the DNA. The affected resonances include the backbone amides of N99, I102, L132, K213, A214 and the side-chain amide of Q134 (Figure 4D and E). Compared to the free-protein spectrum we only miss T211, which cannot be identified. The perturbation induced by the b10 DNA was complete at equimolar concentration with cERCC1. Chemical shift mapping reveals that all perturbations are in the vicinity of the C-terminus of the cERCC1 construct, consistent with the presence of a flexible tag interfering with DNA binding. The DNA-binding region resides at the surface of the structure where the N- and C-termini meet (Figure 4F). The perturbed residues belong to well-defined structure elements. The only positively charged residue we identified (K213) is fully conserved in the ERCC1 proteins and may explain the dependence of DNA binding on salt concentration. All the other affected residues are less conserved than the residues involved in XPA binding. The composition of the affected residues together with the slow exchange behavior suggests a large contribution to the binding by hydrophobic interactions with the DNA bases.
Summarizing, both the biochemical and the NMR experiments show that the ERCC1 central domain binds to DNA. The DNA-binding site we identified on cERCC1 by the NMR titrations suggests that cERCC1 contacts only a small part of the ssDNA, probably three to four bases. The situation is different in the full-length heterodimer, where other DNA-binding surfaces, such as previously established for the C-terminal ERCC1/XPF domains (12,14), assist in DNA recognition.
cERCC1 can bind simultaneously to XPA and ssDNA
Chemical shift perturbation experiments revealed distinct binding surfaces for ssDNA and XPA, and therefore, it is probable that both can bind simultaneously to cERCC1. We performed EMSA experiments in the presence of an equimolar or 5-fold excess of the HIS-XPA peptide. Addition of XPA did not influence binding of cERCC1 to ssDNA, while the binding affinity of XPA for the cERCC1 domain under these conditions would suggest that the vast majority of cERCC1 is in complex with XPA. We did not observe formation of a slower migrating HIS-GST-cERCC1/ssDNA/XPA super complex, possibly because the increase in mass by the addition of the HIS-XPA peptide was too small to discern (data not shown).
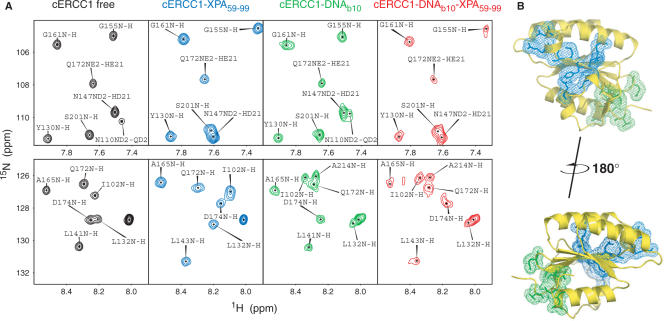
To independently confirm the formation of a ternary cERCC1/ssDNA/XPA complex, we added the HIS-XPA peptide to the cERCC1-bubble10 protein–DNA complex and followed the chemical shift perturbations in the 1H-15N HSQC spectra. As shown in Figure 5, addition of an equimolar amount of the XPA peptide causes the same amide displacements as in the titration with the free protein. On the contrary, most amides influenced by the DNA binding remain unaffected by the presence of XPA (Figure 5A). I102 and N110 were perturbed by both XPA and b10 DNA when the titrations were performed independently. In the ternary complex, however, I102 remains close to the position as in the DNA bound form. Conversely, the side-chain amide of N110, although affected by the DNA, adopts the distinct XPA bound position upon XPA addition. These data strongly indicate the formation of the ternary ERCC1/DNA/XPA complex. DNA and XPA-binding sites are distinct, and the two interactions can happen simultaneously (Figure 5B).
Figure 5.
Dual function of cERCC1. (A) Indicative portions of the 1H-15N HSQC spectrum for the free cERCC1, bound to either XPA (1:1) or DNA (1:1) separately, and the ternary complex (1:1:1). (B) Views of the two binding sites on cERCC1 structure.
Functional role of the ERCC1 central domain in the heterodimer
Since the catalytic activity has been preserved in the human XPF protein through strong conservation of the nuclease signature (5), the human nuclease fold should be identical to that of the archaeal counterparts. In that sense, human XPF nuclease and human ERCC1 central domains are expected to exhibit the same fold. Moreover, the ERCC1 and XPF HhH2 domains feature a common architecture and come into tight association exactly in the same way that the HhH2 domains of both archaeal species do (10,11,14). Therefore, in structural terms the human ERCC1 and XPF proteins share the essential architectural subunits observed in the short XPF homodimer of A. pernix. The structural similarities within the XPF family provide additional evidence for the proposed common origin of the human ERCC1 and XPF proteins (33). Because ERCC1 is absent in archaea, this gene is thought to have been acquired from an ancient XPF gene duplication in the eukaryal lineage. The ERCC1 and XPF genes have subsequently evolved by the process called subfunctionalization (36,37). This model suggests that after gene duplication, both copies may be reciprocally preserved through the fixation of complementary loss-of-subfunction mutations, which results in a partitioning of the tasks of the ancestral gene. From the functions present in the ancestral XPF protein (archaea), human ERCC1 retained the canonical HhH2 domain that acts as a DNA-binding domain (14), while XPF retained the catalytic activity (5). Once the separation of the ancestral subfunctions occurred in ERCC1 and XPF genes, only the heterodimeric protein complex could restore the original function. Additionally, due to genetic alterations, the second helix-hairpin-helix motif of human XPF degenerated, yet the fold remained crucial for stabilizing the corresponding intact domain of human ERCC1 (14). Similarly, the central domain of ERCC1 lost the catalytic activity by sequence drift, but despite adoption of a novel dual function (XPA interaction and DNA binding), the 3D fold was preserved.
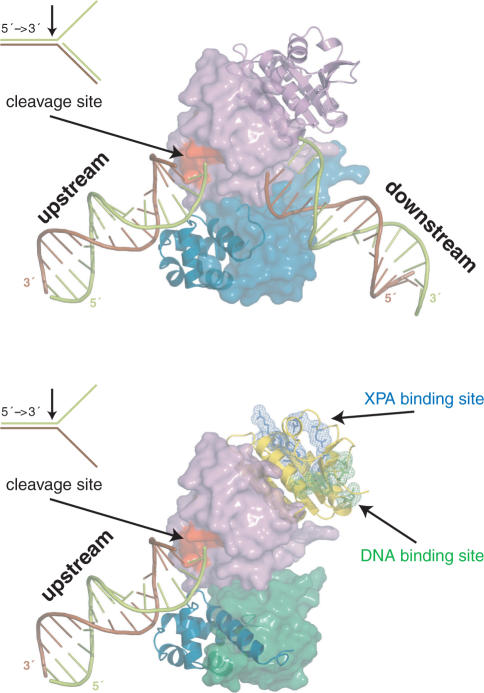
We suggest a mechanistic model for the heterodimeric function based on the model for the homodimeric archaeal homolog (Figure 6) (10,38). Human ERCC1 corresponds to the archaeal protomer that binds with the HhH2 domain to the upstream duplex. Human XPF corresponds to the archaeal protomer that recognizes the downstream duplex. Since it cannot make contacts with the minor groove (14), the specificity of the human heterodimer has shifted to splayed-arms substrates consisting of only one duplex. This does not exclude contribution of the XPF HhH domain to ssDNA interactions, as reported before (12). We show here that the central domain of ERCC1 is also involved in ssDNA binding. Additional DNA interactions are likely for the nuclease of XPF, analogous to the archaeal case (38). Furthermore, ssDNA binding by cERCC1 will be stimulated by specific interactions with XPA, already present in the NER pre-incision complex and possibly bound to the DNA via its own DNA-binding site. Most importantly, through these coordinated interactions of the ERCC1 domains the XPF protein will be positioned to cleave the 3′ protruding strand (limon), thereby retaining the polarity present in the archaeal homodimer.
Figure 6.
Models for the function of the archaeal homodimeric XPF (top) and human ERCC1/XPF heterodimer (bottom) constructed from the structure of the XPF homodimer bound to dsDNA (2bgw), the free structure of ERCC1/XPF C-terminal interacting domains (1z00) and the current structure of the ERCC1 central domain (2jpd). For the homodimer, one protomer is shown in a cartoon and the other in a surface representation. Accordingly, for the heterodimer ERCC1 is in a cartoon and XPF in a surface representation. The protein domains in both cases are colored as in Figure 1A.
Our model underlines the significant role of ERCC1 in the context of the full-length heterodimer. XPF is the catalytic module but the ERCC1 domains guarantee that the enzymatic activity is targeted properly. The presence of multiple distinct DNA-binding surfaces within the ERCC1/XPF/XPA/RPA repair protein intermediate coordinates cleavage to occur only when the DNA damage is recognized correctly by the NER machinery. The duplication of the ancestral XPF gene within the eukaryal kingdom resulted in an obligate heterodimer through loss of function (with an altered HhH2 domain of XPF and a degenerated catalytic domain of ERCC1) and adoption of novel functions (ssDNA and XPA binding of cERCC1). This permitted additional quality control mechanisms through a more complicated molecular interaction network, mediated by the novel functional domains, and thereby improving the fidelity of DNA damage repair.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank Koos Jaspers and Jan Hoeijmakers for providing the full-length vectors of ERCC1 and XPF, and for many fruitful discussions. This work was financially supported by the Netherlands Organization for Scientific Research, Chemistry Council (NWO-CW), by the Center of Biomedical Genetics, the Netherlands, and by the EU project Spine2-Complexes (Contract no 031220). Funding to pay the Open Access publication charges for this article was provided by NWO-CW.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nishino T, Morikawa K. Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors. Oncogene. 2002;21:9022–9032. doi: 10.1038/sj.onc.1206135. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Nishino T, Komori K, Ishino Y, Morikawa K. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: similarity between its endonuclease domain and restriction enzymes. Structure (Camb.) 2003;11:445–457. doi: 10.1016/s0969-2126(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 4.Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem. Sci. 2003;28:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Enzlin JH, Scharer OD. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NGJ, Beverloo HB, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JHJ, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 8.Jaspers NGJ, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, Giglia-Mari G, Hoogstraten D, Kleijer WJ. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am. J. Hum. Genet. 2007;80:457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 10.Newman M, Murray-Rust J, Lally J, Rudolf J, Fadden A, Knowles PP, White MF, McDonald NQ. Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J. 2005;24:895–905. doi: 10.1038/sj.emboj.7600581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishino T, Komori K, Ishino Y, Morikawa K. Structural and functional analyses of an archaeal XPF/Rad1/Mus81 nuclease: asymmetric DNA binding and cleavage mechanisms. Structure. 2005;13:1183–1192. doi: 10.1016/j.str.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 12.Tsodikov OV, Enzlin JH, Scharer OD, Ellenberger T. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc. Natl Acad. Sci. USA. 2005;102:11236–11241. doi: 10.1073/pnas.0504341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Laat WL, Appeldoorn E, Jaspers NGJ, Hoeijmakers JHJ. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- 14.Tripsianes K, Folkers GE, AB E, Das D, Odijk H, Jaspers NGJ, Hoeijmakers JHJ, Kaptein R, Boelens R. The structure of the human ERCC1/XPF interaction domains reveals a complementary role for the two proteins in nucleotide excision repair. Structure. 2005;13:1849–1858. doi: 10.1016/j.str.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 16.Missura M, Buterin T, Hindges R, Hubscher U, Kasparkova J, Brabec V, Naegeli H. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–3564. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JHJ, van Driel R, van Zeeland AA, et al. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Peterson CA, Lu X, Legerski RJ. Mutations in XPA that prevent association with ERCC1 are defective in nucleotide excision repair. Mol. Cell. Biol. 1995;15:1993–1998. doi: 10.1128/mcb.15.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Jong RN, Daniels MA, Kaptein R, Folkers GE. Enzyme free cloning for high throughput gene cloning and expression. J. Struct. Funct. Genomics. 2007 doi: 10.1007/s10969-006-9014-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Folkers GE, van Buuren BN, Kaptein R. Expression screening, protein purification and NMR analysis of human protein domains for structural genomics. J. Struct. Funct. Genomics. 2004;5:119–131. doi: 10.1023/B:JSFG.0000029200.66197.0c. [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2001. [Google Scholar]
- 24.Cavanagh J, Fairbrother JW, Palmer GAIII, Skelton JN. Protein NMR Spectroscopy. San Diego, CA, USA: Academic Press; 1996. [Google Scholar]
- 25.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 26.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 27.Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CAEM, Nabuurs SB, Güntert P, Livny M, Markley JL, et al. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–762. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 28.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta. Crystallogr. D Biol. Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.DeLano W. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 30.Singh S, Folkers GE, Bonvin AMJJ, Boelens R, Wechselberger R, Niztayev A, Kaptein R. Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli. EMBO J. 2002;21:6257–6266. doi: 10.1093/emboj/cdf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonker HR, Wechselberger RW, Boelens R, Folkers GE, Kaptein R. Structural properties of the promiscuous VP16 activation domain. Biochemistry. 2005;44:827–839. doi: 10.1021/bi0482912. [DOI] [PubMed] [Google Scholar]
- 32.Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 33.Gaillard PH, Wood RD. Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res. 2001;29:872–879. doi: 10.1093/nar/29.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Elledge SJ, Peterson CA, Bales ES, Legerski RJ. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl Acad. Sci. USA. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchko GW, Isern NG, Spicer LD, Kennedy MA. Human nucleotide excision repair protein XPA: NMR spectroscopic studies of an XPA fragment containing the ERCC1-binding region and the minimal DNA-binding domain (M59-F219) Mutat. Res. 2001;486:1–10. doi: 10.1016/s0921-8777(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 36.Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynch M, Conery JS. The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics. 2003;3:35–44. [PubMed] [Google Scholar]
- 38.Roberts JA, White MF. An archaeal endonuclease displays key properties of both eukaryal XPF-ERCC1 and Mus81. J. Biol. Chem. 2005;280:5924–5928. doi: 10.1074/jbc.M412766200. [DOI] [PubMed] [Google Scholar]
- 39.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.