Abstract
Active Activator (Ac) elements undergo mutations to become nonautonomous Dissociation (Ds) elements at a low frequency. To understand the mechanism of Ds formation, we have developed high-throughput genetic and molecular screens to identify these rare Ds derivatives generated from any Ac insertion in the maize genome. Using these methods we have identified 15 new Ds elements derived from Ac insertions at eight different loci. Approximately half of the Ds elements contain filler DNA inserted at the deletion junction that is derived from sequences within or adjacent to Ac. In contrast to previous reports, several of these Ds elements lack direct repeats flanking the deletion junctions and filler DNA in the donor Ac. To accommodate our findings and those of others, we propose a model of slip mispairing during error-prone repair synthesis to explain the formation of state II Ds elements in maize. We discuss the use of these lines and molecular techniques developed here to capture somatic Ds transposition events in two-component Ac/Ds tagging programs in maize.
THE Activator/Dissociation (Ac/Ds) transposon family has been extensively characterized since its discovery in maize >60 years ago (McClintock 1946; Kunze and Weil 2002). Ac/Ds are class II DNA transposons that belong to the hAT superfamily of plant transposable elements (Kunze and Weil 2002). Ac is a 4565-bp autonomous element capable of catalyzing the transposition of itself and nonautonomous Ds elements (McClintock 1949, 1951). Ac encodes a 3.5-kb open reading frame (ORFa) that directs the synthesis of an 807-amino-acid transposase (TPase) essential for both Ac and Ds transposition (Fedoroff et al. 1983; Kunze et al. 1987). The 11 bp imperfect terminal inverted repeats (TIR) and ∼240 bp of subterminal sequences are critical for TPase binding and transposition of both Ac and Ds (Coupland et al. 1988, 1989).
In contrast to the highly conserved structure of Ac elements (Fedoroff et al. 1983; Behrens et al. 1984; Muller-Neumann et al. 1984; Pohlman et al. 1984), Ds elements are structurally diverse. State I or “double Ds” elements consist of nested Ds insertions or contain multiple Ds end sequences in close proximity (Doring et al. 1984, 1989, 1990; Weck et al. 1984; Ralston et al. 1989; Weil and Wessler 1993). State II elements are likely deletion derivatives of Ac that differ in size and internal structure (reviewed in Kunze and Weil 2002). For instance, Ds1-like elements contain large internal deletions sharing only the 5′ terminal 13 bp and 3′ terminal 26 bp of sequence with Ac (Sutton et al. 1984). The Ds9 element is identical to Ac with the exception of a 194-bp simple deletion in the element (Rubin and Levy 1997). Other Ds family members, such as wxB4, share the TIR and subterminal sequence with Ac but carry internal sequences with no homology to Ac (Varagona and Wessler 1990). State I elements often induce chromosome breakage but rarely transpose whereas state II elements transpose frequently but rarely break chromosomes (McClintock 1946, 1949).
Ac and Ds have been used to clone and characterize many genes in maize (for reviews see Kunze et al. 1997; Brutnell 2002; Settles 2005). Strategies to utilize these elements in gene-tagging programs have been devised that exploit the tendency of Ac and Ds to transpose to closely linked sites (Van Schaik and Brink 1959; Greenblatt 1984; Dooner and Belachew 1989). To facilitate the use of Ac in tagging programs, several Ac elements have been mapped throughout the maize genome (Auger and Sheridan 1999; Cowperthwaite et al. 2002), including 60 that have been precisely positioned on genetic and physical maps (Singh et al. 2003; Kolkman et al. 2005).
Although Ds has not been used extensively for gene tagging in maize (Kunze et al. 1997; Brutnell and Conrad 2003), it does afford several advantages over Ac. Foremost, Ds-induced mutations allow for the analysis of either stable or mutable phenotypes. In the absence of an Ac transposase source, Ds insertions are stable, facilitating physiological studies that would be complicated by somatic excision. Stable Ds alleles can also be incorporated into breeding programs without the risk of losing the desired allele due to transposon excision. When mutable phenotypes are desirable, for instance, in clonal analysis (Emerson 1917; Dawe and Freeling 1990) or to generate an allelic series (Wessler et al. 1986; Moreno et al. 1992; Weil et al. 1992; Singh et al. 2003), the elements can be remobilized by introducing an Ac element into the genome through crossing. Unfortunately, relatively few Ds insertions have been genetically or physically mapped in the maize genome, limiting the potential of Ds in directed or regional tagging programs (Conrad and Brutnell 2005).
The generation of a two-element system from a one-element system was first observed by McClintock when she identified Ds derivatives from Ac insertions at the Bronze1 and Waxy1 loci of maize (McClintock 1955, 1956, 1962, 1963). More recently, Yan et al. (1999) described the formation of three Ds elements from the bz1-m2∷Ac allele and one from an Ac element inserted 0.05 cM proximal to the Bronze1 locus, designated tac2094∷Ac. Two related mechanisms have been proposed to account for the formation of Ds elements from Ac. Rubin and Levy (1997) concluded that state II Ds elements are generated through an abortive gap-repair process following Ac excision. However, in some instances, a synthesis-dependent strand-annealing (SDSA) repair mechanism is initiated at the time of this abortive gap repair. In these instances, polymerase slippage during DNA synthesis at the site of direct repeats within Ac may result in the insertion of sequences known as filler DNA at the deletion junction (Rubin and Levy 1997). Yan et al. (1999) proposed that Ds elements were formed through slip mispairing during DNA synthesis resulting both in an internal deletion and in some cases the insertion filler DNA at the deletion junction (Yan et al. 1999). Both models propose that slip mispairing of direct repeats at the deletion breakpoints within Ac occurs during DNA synthesis.
In addition to Ac, the formation of nonautonomous elements via double-strand break (DSB) repair has been studied for several class II transposon families such as Mutator in maize (Hsia and Schnable 1996), P-elements in Drosophila (O'hare and Rubin 1983; Takasu-Ishikawa et al. 1992), and Tam3 in Antirrhinum (Yamashita et al. 1999). Although the formation of these nonautonomous elements is thought to occur through similar DNA repair mechanisms, there are differences in the frequency that repair products arise in each of these transposon families (Yamashita et al. 1999). In contrast to P, Mu, and Ac elements, the Tam3 family of elements is extremely conserved in structure due to a lack of the formation of repair products following excision. Yamashita et al. (1999) suggest that hairpin formation at the ends of Tam3 prevents gap repair from proceeding into the element, precluding the formation of functional nonautonomous Tam3 elements. Although it has been shown that both Mu and Ac ends are capable of forming hairpin structures, these elements still produce nonautonomous derivatives (Yamashita et al. 1999). These studies suggest there are likely several factors that affect the DSB repair mechanism following excision of an active transposon including secondary structure.
In this report we describe the development of high-throughput genetic and molecular screens to identify newly formed Ds elements from previously positioned Ac insertions. Using these methods we identified 15 new Ds elements derived from Ac insertions at eight different sites in the maize genome. Sequence analysis revealed significant differences between these 15 Ds elements and previously reported Ds derivatives (Rubin and Levy 1997; Yan et al. 1999). Specifically, several of these Ds elements lack direct repeats flanking the deletion junctions in the donor Ac. Approximately half of the Ds elements contain filler DNA inserted at the deletion junction and this filler DNA often, but not always, originated from within Ac. Thus, we propose a modified version of a model proposed by Dooner and colleagues (Yan et al. 1999) that accounts for the formation of all state II derivatives characterized to date. To further exploit these lines in mutagenesis programs, we developed a “Ds-casting” procedure to capture local somatic Ds transposition events and we discuss the use of these lines in two-component Ac/Ds tagging programs in maize.
MATERIALS AND METHODS
Description of maize stocks:
All maize stocks used in this study were in the W22 inbred background. All Ac lines, with the exception of bti97156∷Ac, were previously described (Kolkman et al. 2005). The bti97156∷Ac line contains an Ac insertion 4 cM from the Pink Scutellum1 (Ps1) gene and has been previously described (Singh et al. 2003). The r1-sc:m3 Ds tester line contains a Ds6-like insertion in the r1 locus that controls anthocyanin accumulation in the aleurone and scutellar tissues (Alleman and Kermicle 1993). In the absence of Ac transposase, this insertion renders the kernels colorless. In the presence of Ac transposase, excision of the Ds from the r1 locus can be visualized as purple sectors in the aleurone. Increasing copy numbers of Ac in the genome result in fewer and smaller sectors known as the negative dosage effect (McClintock 1951). The Ac-immobilized (Ac-im) stock contains an Ac derivative inserted at 7.02, which is incapable of transposition but still encodes a functional transposase protein (Conrad and Brutnell 2005).
Selection for Ds derivatives:
Ac activity was monitored using the r1-sc:m3 Ds tester line described above. Kernels homozygous for an Ac insertion were selected on the basis of the canonical negative dosage effect of Ac (McClintock 1951). Kernels were sown in the field and mature plants were testcrossed to the r1-sc:m3 Ds tester line. Colorless kernels resulting from a loss of Ac activity were selected from the progeny ears and plants self-pollinated to generate segregating families. The resulting ears were screened using one of two methods. When the ps1-m8∷Ac allele was used as a donor Ac, colorless progeny ears were screened visually for segregation of a ps1 mutant phenotype with a lack of Ac activity (Singh et al. 2003; Bai et al. 2007). Progeny ears from all other donor Ac elements were screened using a molecular assay. Ten colorless kernels from self-pollinated F2 ears were sown in greenhouse sandbenches. DNA was extracted from seedling leaf tissue and a verification PCR assay was performed on DNA pools as described below.
DNA extraction:
DNA pools were constructed from ∼1 g of seedling leaf tissue that was collected in two-dimensional pools. Seed from each segregating ear was planted in 10-kernel rows. Leaf tissue from these 10 seedlings were pooled and placed in both a row pool, consisting of 10 rows, and a column pool, consisting of one column from each of the 10 rows. Thus, a unique address could be assigned to each of the 100 segregating families using 20 pooled DNA extractions. Positive results were confirmed by extracting DNA from individual family members and repeating the PCR assay. DNA extraction was performed as described by Chen and Dellaporta (1994).
DNA extractions for the Ds-casting assay (see below) were conducted in a 96-well format. Two centimeters of leaf tissue were collected from the base of the youngest leaf of field-grown plants ∼50 days after planting. The midrib was removed and tissue was placed in a 96-well plate with two ball bearings. Tissue was frozen in liquid nitrogen and pulverized in a paint shaker for 3 min. A phenol-chloroform extraction was performed as previously described (Chen and Dellaporta 1994).
PCR amplification:
Verification PCR:
PCR was performed on pooled DNA extractions described above. PCR primers designed for Ac insertion verification described in Kolkman et al. (2005) were utilized to amplify a region from the DNA flanking the element into the end of putative Ds derivatives (supplemental Table 1 at http://www.genetics.org/supplemental/). Verification PCR was performed using ∼6 ng of total genomic DNA in a 50 μl total reaction volume according to reaction conditions described in Conrad and Brutnell (2005). PCR products were fractionated on a 0.8% agarose gel containing ethidium bromide and visualized on a UV light box.
Amplification of Ds elements:
The long-range inverse PCR (IPCR-3) protocol described in Kolkman et al. (2005) was used to amplify deletion breakpoints from the Ds derivatives. PCR was performed on ∼6 ng of genomic DNA extracted from five pooled seedling leaf tissue samples from individual segregating ears. Flanking primers presented in supplemental Table 1 at http://www.genetics.org/supplemental/ were used in conjunction with Ac primers. For the ps1-m8∷Ds alleles D1, D4, and D5, sequences were amplified with the Ps1-21 primer (supplemental Table 1) while the ps1-m8∷Ds(D3) allele was amplified with the Ps1-10 primer described in Singh et al. (2003). The ps1-m8∷Ds(D2) was amplified with two flanking primers Ps1-21 (supplemental Table 1) and Ps1-10 (Singh et al. 2003). Ac primers used to amplify toward the 5′ Ac flanking sequences were: 191.Ac4: 5′-CAATAGCCATATCATCTTGACTCG, TBp34, Ac14, and TBp41 reported in (Kolkman et al. 2005). Ac primers used to amplify toward the 3′ Ac flanking sequences were: TEB252.35Ac1: 5′-CAGGGATGAAAGTAGGATGGGAAA, TEB252.35Ac3: 5′-CAAGCTGTTGTGTCATTTGTGTGC, Ac2: 5′-TTCATGTGAGGTGTGCTTGTC, 191.Ac5: 5′-GTGCTAGACTCTGTTATTGCTGCT, TBp38, Tbp40, and JGp2 reported in Kolkman et al. (2005). PCR products were fractionated on a 0.8% agarose gel with ethidium bromide and visualized on a UV light box.
Ds casting:
A line homozygous for the bti00252∷Ds(D2) derivative was used to pollinate a line homozygous for the Ac-im insertion. The resulting progeny were planted in the field and seedling leaf tissue was collected. DNA extractions were performed according to the protocol described above and a Ds-casting protocol was adapted from Singh et al. (2003). Two rounds of nested PCR were performed using the IPCR-3 reaction protocol described in Kolkman et al. (2005) with nested flanking and Ds end primers (supplemental Figure 2A at http://www.genetics.org/supplemental/). The first round of PCR used primers bti00252A.F2: 5′-CTAAGCTGACTGTTGTGTGGGAAG and Ac3: 5′-CATATTGCAGTCATCCCGAA with ∼1–3 μg of genomic DNA in a total reaction volume of 20 μl. First round products were diluted 1:200 in dH2O and 1 μl of this dilution was used as a template for the second round of nested PCR with primers bti00252A.F7: 5′-CGTACGTGTCATAACTTTTGGAAG and TBp35 (described in Kolkman et al. 2005). PCR products were fractionated on a 1% agarose gel with ethidium bromide and visualized on a UV light box (supplemental Figure 2B).
Sequencing:
PCR products were separated on a 0.8% agarose gel and purified using the QIAquick gel extraction kit (QIAGEN, Valencia, CA). DNA was cloned into either pGEM-T Easy (Promega, Madison, WI) or the TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA) and sequenced as previously described (Singh et al. 2003).
RESULTS
Genetic selection for newly formed Ds derivatives:
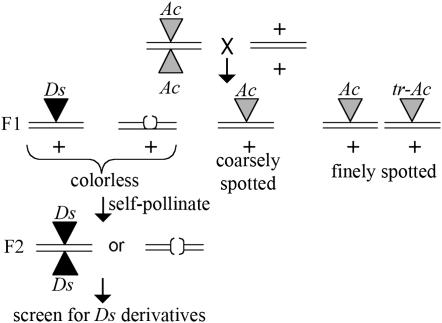
Ac donor lines were selected from a subset of previously characterized insertions (Kolkman et al. 2005) and from an Ac insertion at the pink scutellum1 (ps1) locus, ps1-m8∷Ac, which conditions a weak mutant phenotype when homozygous (Bai et al. 2007). Ac activity was visualized as purple sectors on the kernels resulting from excision of a Ds element from the r1 locus during kernel development (r1-sc:m3; see materials and methods). Thus, a loss of Ac activity was scored as the lack of these purple sectors. To identify deletion derivatives of Ac, lines homozygous for an Ac insertion and the r1-sc:m3 allele were testcrossed by the Ds tester line (Figure 1). The majority of progeny kernels from F1-testcross progeny are coarsely spotted as a consequence of being hemizygous for the original Ac insertion. Approximately 2–4% of the progeny were finely spotted resulting from an increase in Ac copy number due to a new transposition (tr-Ac) (Kolkman et al. 2005). Rare colorless kernels were selected, grown, and self-pollinated to identify ears that were completely colorless in the F2 generation due to a loss of Ac activity. Consequently, Ds derivative events that cosegregate with a tr-Ac in the genome would not be detected in our screen.
Figure 1.—
Genetic selection for newly formed Ds derivatives. Lines homozygous for an Ac insertion (shaded triangles) were pollinated by the Ds tester line. All lines are homozygous for the r1-sc:m3 allele (not shown). Colorless kernels either containing a new Ds (solid triangles) or a loss of Ac due to excision (parentheses) were selected from the resulting progeny ears. Colorless kernels were sown and mature plants were self-pollinated resulting in an ear segregating for either a new Ds or the Ac excision site. Ears derived from the ps1-m8∷Ac lines were visually screened for a mutant phenotype. Kernels from the remaining ears were planted and DNA was extracted from seedling leaf tissue. PCR was used to screen for the insertion of putative Ds at the original locus as described in the text.
Approximately 0.6% of the F1-testcross progeny kernels were colorless due to a loss of Ac activity. It is likely that in the majority of these cases, the loss of Ac activity was due to premeiotic excision of Ac followed by an independent assortment of the donor locus and transposed Ac (Greenblatt 1984). In rare cases, Ac will undergo a mutational event that renders it inactive. Both losses of Ac from the genome and putative inactivation events are selected in the genetic scheme outlined in Figure 1. A total of 753 colorless progeny ears were screened representing colorless kernel derivatives from 20 loci containing Ac insertions, including the ps1-m8∷Ac allele. As described in the following sections, 24 Ac inactivation events, or putative Ds elements, were identified at 8 loci, including 6 from the ps1-m8∷Ac allele. These putative Ds insertions were further characterized through phenotypic or PCR-based screens to confirm the presence of an insertion at the original locus as detailed below. Importantly, this genetic screen could be applied to detect Ds derivatives from any Ac insertion in the maize genome.
Identification of Ds insertions through a molecular screen:
We developed a general PCR assay to distinguish putative Ds derivatives from Ac excision events. As shown in Figure 1, putative Ds derivatives would be expected to segregate in self-pollinated F2 families. Thus, 10 kernels from colorless F2 ears were planted in the greenhouse and DNA was extracted from pooled seedling leaf tissue samples (materials and methods). PCR primers were designed to amplify a fragment immediately adjacent to the original Ac insertion using a subterminal Ac end- and flanking sequence-specific primer pair (supplemental Table 1 at http://www.genetics.org/supplemental/). Germinal excisions of Ac would not result in amplification products whereas those elements that retained the sequences complementary to the primer at the original insertion site would result in an amplification product of a predicted size. We screened 673 families and identified 18 that contained an Ac-end sequence at the site of the original insertions but lacked Ac activity. These families were selected as carrying putative novel Ds insertions.
One limitation of this screen was the requirement for intact Ac end sequences at the priming site. In other words, if a more complex rearrangement of Ac sequences resulted in the deletion of end sequences containing the primer sequences, the event would not be detected in the screen. Nevertheless, the screen does enrich for events that are likely to generate a functional Ds derivative.
Identification of Ds insertions through a phenotypic screen:
To investigate the diversity of Ac-mediated deletions and more complex rearrangements, we utilized an Ac insertion at the ps1 locus (ps1-m8∷Ac). In contrast to the molecular screen described above, this phenotypic screen did not require intact Ac ends or flanking sequences for the identification of a Ds derivative at this locus. Ps1 encodes an enzyme with lycopene β-cyclase activity that catalyses the first committed step in xanthophyll biosynthesis (Singh et al. 2003). Although the majority of mutations in Ps1 are lethal, ps1-m8∷Ac kernels germinate at nearly the same frequency as wild type (Bai et al. 2007). Seedlings homozygous for the ps1-m8∷Ac allele have slightly pale green leaf sheath tissue and virescent leaf blade tissue and kernels from self-pollinated ears are slightly pink due to an accumulation of δ-carotene in the endosperm (L. Bai, unpublished results; Bai et al. 2007). We reasoned that Ds derivatives would condition for a similar mutant phenotype as the donor Ac insertion and thus could be identified as slightly pink kernels lacking Ac activity.
Genetic selections were performed as described above and colorless F2 ears were visually screened for segregation of a weak ps1 mutant phenotype segregating in a 3:1 Mendelian ratio (Singh et al. 2003; Bai et al. 2007). Six alleles were selected as having a weak mutant phenotype and lacking Ac activity. Ds derivatives that restore wild-type Ps1 function would not be detected in this screen.
Ds deletion junction sequence analysis:
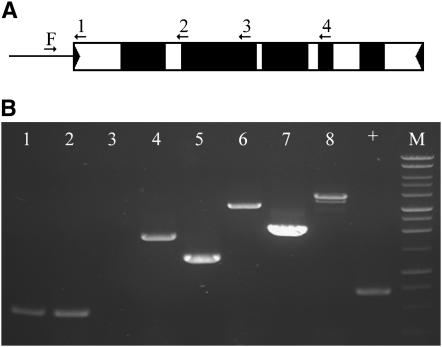
To define the deletion breakpoints in each of the putative Ds derivatives, a long-distance PCR method was utilized to rapidly survey the structure of the element (materials and methods). As illustrated in Figure 2A, PCR primers designed to sequences throughout the Ac element were used in conjunction with an Ac-flanking primer as described by Kolkman et al. (2005). As shown in Figure 2B, if the primer sequence is contained in the deleted portion of the Ds, a PCR product will not be generated (lane 3) while primers flanking this deletion will give a smaller product (lanes 5 and 7) when compared to a full-length Ac (lanes 6 and 8). PCR products containing the deletion breakpoint were then sequenced (see materials and methods). This PCR assay enabled the rapid identification of the sequences deleted from 15 of the 24 putative Ds elements.
Figure 2.—
Amplification across Ds deletion junctions. (A) Ac schematic. Flanking DNA upstream of the Ac/Ds represented by a line and Ac sequence shown as rectangle with TIRs as solid triangles, exons as solid boxes, and noncoding seqeunces as open boxes. Primers are shown as arrows, F is the flanking primer, and 1–4 are internal Ac primers. (B) Ethidium bromide-stained agarose gel showing PCR amplification products derived from a new Ds element. Lanes 2, 4, 6, and 8 show PCR products generated using bti97156∷Ac as a template and lanes 1, 3, 5, and 7 show PCR products derived from bti97156∷Ds as a template; the + is a positive control, and the M is a Promega 1-kb ladder. Primer F was used in all reactions: lanes 1 and 2, F + primer 1; lanes 3 and 4, F + primer 2; lanes 5 and 6, F + primer 3; lanes 7 and 8, F + primer 4.
The nine derivatives that did not appear to result from simple deletions of Ac were further characterized. Four contained an apparently full-length Ac element inserted at the original locus, based on PCR product size even though no Ac activity was detected. There are at least two possibilities that could account for the loss of Ac activity in lines that contain an apparently full-length Ac. First, small deletions (≤50 bp) would not be detected in the PCR assays, but still disrupt Ac activity. Alternatively, epigenetic silencing associated with increased methylation of internal Ac sequences (Schwartz and Dennis 1986; Chomet et al. 1987; Kunze et al. 1987, 1988; Fusswinkel et al. 1991; Brutnell and Dellaporta 1994; Brutnell et al. 1997) may have resulted in the loss of Ac activity. To examine this latter possibility, DNA gel blot analysis was performed on families that contained an apparently full-length Ac element at the original locus. To monitor the methylation status of diagnostic nucleotides within Ac, DNA was digested with the methylation-sensitive restriction enzyme PvuII and blots were hybridized with the internal Ac900 or Ac700 fragments described in Kolkman et al. (2005). This analysis suggests at least one of the putative Ds elements was methylated at the internal PvuII sites (data not shown), indicating that the loss in activity in some families was due to epigenetic modification of Ac sequences. The methylation of the internal PvuII sites does not appear to be the consequence of Ds formation, as derivative bti00228∷Ds retained two unmethylated PvuII sites. The five remaining putative Ds elements retained Ac end sequences at the original locus but we were unable to amplify products in the long-distance PCR assay, despite repeated attempts. These insertions may have resulted from more complex rearrangements of Ac sequences rather than simple deletions. DNA blot analysis of three of these derivatives indeed suggests a more complex rearrangement of sequences (data not shown).
Frequency of Ds formation:
A total of 15 new Ds elements were identified from eight unique Ac insertions in the maize genome. Table 1 presents the minimum frequency of Ds formation for several of the Ac-containing lines screened. Accurate progeny counts were only obtained for four Ac donor lines and of these, screens of only one line (bti00252∷Ac) resulted in the recovery of multiple Ds insertions. In this instance, the frequency of Ds formation was calculated at 0.03%. However, if we consider the total number of kernels screened regardless of Ac donor, we estimate the frequency of Ds formation at 0.025% (5/19,923 × 100).
TABLE 1.
Frequency of Ds formation
| Ac progenitor | Kernels screened | Ds identified |
|---|---|---|
| mon03077∷Ac | 9965 | 1 |
| bti00228∷Ac | 895 | 1 |
| bti00252∷Ac | 6948 | 2 |
| bti00257∷Ac | ND | 3 |
| 1.07∷Ac | ND | 1 |
| bti00191∷Ac | 2115 | 1 |
| bti97156∷Ac | ND | 1 |
| ps1-m8∷Ac | ND | 5 |
Ds elements contain novel deletions:
As predicted from previously published studies (Rubin and Levy 1997; Yan et al. 1999), a preferential size or location of deleted Ac sequence was not observed (Figure 3). The size of deletions ranged from 388 bp in bti00191∷Ds to 3505 bp in ps1-m8∷Ds(D2) and were not restricted to any particular region within Ac (Table 2, Figure 3). Interestingly, bti00257∷Ds(D1) carries an in-frame deletion of amino acid residues 331–475 of exon 2. This derivative is associated with a tightly linked Ac insertion but conditions a finely spotted aleurone that appeared nearly colorless in initial kernel selections. This aberrant spotting pattern suggests that bti00257∷Ds(D1) encodes a protein that interacts with Ac and alters its activity. We have not yet separated the tightly linked Ac element from this derivative and thus, have not further characterized this element in this study. None of the other derivatives encoded proteins with any detectable activity.
Figure 3.—
Structures of Ds derivatives. The internal structures of 15 Ds derivatives characterized in this study are shown in comparison to Ac (top). Solid lines represent noncoding sequences and solid rectangles are exons. The arrowheads denote the start of transcription. Deletions within Ds elements are shown as a gap. Filler DNA is denoted by shaded boxes and the origin of the filler DNA sequence, when known, is shown as open boxes.
TABLE 2.
Deletion derivatives identified in this study
| Map position | Derivative | Deletion size (bp) | Presence of filler DNA (bp) | Origin of filler DNA |
|---|---|---|---|---|
| 1.01 | mon03077∷Ds | 1553 | None | NA |
| 1.03 | bti00228∷Ds | 710 | None | NA |
| 1.04/1.05 | bti00252∷Ds(D1) | 1274 | 29 | Ac |
| bti00252∷Ds(D2) | 1345 | 33 | Ac | |
| 1.05 | bti00257∷Ds(D1) | 435 | None | NA |
| bti00257∷Ds(D2) | 1848 | None | NA | |
| bti00257∷Ds(D3) | 1120 | 4 | Ac | |
| 1.07 | 1.07∷Ds | 673 | 10 | Ac |
| 2.02 | bti00191∷Ds | 388 | None | NA |
| 5.04 | bti97156∷Ds(D1) | 1806 | None | NA |
| 5.04 | ps1-m8∷Ds(D1) | 1141 | 38 | Ac |
| ps1-m8∷Ds(D2) | 3505 | 53 + 11 | Ps1 + Aca | |
| ps1-m8∷Ds(D3) | 815 | 51 | Ac | |
| ps1-m8∷Ds(D4) | 465 | None | NA | |
| ps1-m8∷Ds(D5) | 1815 | None | NA |
Fifty-three base pairs of filler DNA originating from flanking DNA and 11 bp originating in the Ac sequence.
Seven of the 15 Ds derivatives have filler DNA inserted at the deletion junction (Table 2). Filler DNA refers to extraneous nucleotides that are copied into sequence rearrangements from elsewhere in the genome. It is a common result of DSB repair and has been observed in plants, animals, and fungi (for example, see Roth et al. 1989; Sainsard-Chanet and Begel 1990; Wessler et al. 1990; Bundock and Hooykaas 1996; Puchta 2005). The size of the filler DNA varied from 1 to 64 bp and was always found at the deletion junctions. All of the filler DNA originated from sequence in close proximity to the deletion breakpoint and the majority of it is present elsewhere within the Ds (Figure 3). The one exception, ps1-m8∷Ds(D2), contains filler DNA that originated from the flanking sequence adjacent to the site of Ac insertion at ps1 and is missing one Ds end sequence. The derivatives ps1-m8∷Ds(D4) and bti00257∷Ds(D3) have 1 and 4 bp of filler DNA, respectively, inserted at the deletion junction. As the sequence of the filler DNA occurs multiple times within Ac, it precludes determination of the template DNA. Four of the 7 Ds derivatives contain filler DNA that originated from multiple locations and is assembled at the deletion junction (Figure 4 and supplemental Figure 1 at http://www.genetics.org/supplemental/). Interestingly, these filler sequences are not always arranged at the deletion junction in the same order as they occur in their original context in Ac. The most extreme example of this is the ps1-m8∷Ds(D3) element where there are five tandem pieces of filler DNA inserted at the deletion junction plus two additional nucleotides. This mosaic of filler DNA originates from upstream, downstream, and from within the deleted sequences of the Ds with the same 8 bp of filler sequence being repeated twice at the deletion junction (supplemental Figure 1). In summary, to the extent the filler origin can be determined, it always originates from sequence in close proximity to the deletion, can arise from within or outside the Ac sequence, and may be assembled into tandem arrays.
Figure 4.—
Sequence at Ds deletion junctions. Two general classes of Ds elements are defined. Type I are defined by the presence of direct repeats of Ac sequence at the junction of the Ds breakpoint or filler DNA. Type II elements are defined by the absence of direct repeats of Ac sequence at the junction of the Ds breakpoint or filler DNA. Sequence alignment for an example of each Ds type is shown. A single strand of Ac sequence (top) and Ds (bottom) is shown. Numbering is based on published Ac sequence (GenBank accession no. X05424). Direct repeats are surrounded by boxes.
The current model for the formation of Ds derivatives including the insertion of the filler DNA into the deletion junction is based on the observation that short direct repeats flank both the deletion and the origin of filler DNA (Yan et al. 1999). However, in this study, several Ds derivatives do not have direct repeats flanking the deletion junction and/or filler DNA in the Ac donor (Figure 4). We classified the Ds derivatives reported here into two types with regard to the presence or absence of these direct repeats. An example of each type is presented in Figure 4. Type I derivatives (8/15) contain direct repeats in the donor Ac flanking the deletion junction and filler DNA as predicted by previous models. Type II derivatives (5/15) do not contain direct repeats flanking either the deletion junction or, when present, the filler DNA. As mentioned above, two derivatives, ps1-m8∷Ds(D4) and bti00257∷Ds(D3), contain 1 and 4 bp of filler DNA, respectively, so it is not possible to classify them as either type I or type II. Thus, at least 33% of the deletions identified lack direct repeats flanking filler DNA or deletion breakpoints in the donor Ac, indicating that repeat sequences at deletion/filler breakpoints are not prerequisites for state II Ds element formation in maize.
Ds elements capable of somatic transposition:
With the exception of ps1-m8∷Ds(D2), all of the Ds derivatives reported here contain internal deletions and are likely to be transposition competent (Coupland et al. 1988, 1989). To test this prediction and extend the utility of these Ds insertions in gene-tagging programs, we developed a PCR-based method to capture somatic transpositions of Ds. This method was modified from the “Ac-casting” procedure described by Singh et al. (2003). To demonstrate this method, a plant carrying the Ds insertion bti00252∷Ds(D2) was crossed by an individual homozygous for Ac-im (materials and methods). Ac-im is a stabilized source of transposase, capable of mediating somatic and germinal Ds excisions (Conrad and Brutnell 2005). PCR primers were designed to sequences flanking the Ds insertion (GenBank accession no. AY559233) and the 5′-Ds end (supplemental Figure 2A at http://www.genetics.org/supplemental/) to amplify short-range somatic Ds transpositions. Amplification products are expected only when the Ds element excises and reinserts upstream of the flanking primer in the opposite orientation as the original insertion. The results of Ds casting are shown in supplemental Figure 2. Multiple PCR products were detected and sequenced. Some of these products did not contain bti00252 flanking DNA and were likely the result of Ds–Ds amplification products generated from cryptic Ac-like elements present elsewhere in the maize genome. However, one product was recovered that was the result of a local somatic transposition from bti00252∷Ds(D2) (supplemental Figure 2B). Sequence analysis of this fragment revealed that the Ds element excised from the original insertion site generating an 8 bp excision allele and reinserted in the opposition orientation 282 bp upstream of the original insertion. These results demonstrate Ds casting as a method to obtain additional genomic sequences flanking Ds elements in the genome and shows that bti00252∷Ds(D2) is transposition competent.
DISCUSSION
Development of two-component Ac/Ds tagging systems:
Ac and Ds offer a number of advantages for gene-tagging experiments in maize. Both elements are present at relatively low copy number in the genome facilitating molecular and genetic screens (Fedoroff et al. 1983). They display a high frequency of somatic excision that can be exploited to confirm the identity of a tagged gene (Schultes et al. 1996; Schauser et al. 1999), to create an allelic series (Athma et al. 1992; Moreno et al. 1992), or to generate stable “footprint” alleles (Weil et al. 1992; Alleman and Kermicle 1993; Bai et al. 2007). Of particular value are small in-frame insertions following Ac/Ds excision that subtly alter the protein activity (Wessler et al. 1986; Alleman and Kermicle 1993; Giroux et al. 1996; Bai et al. 2007). Finally, both Ac and Ds display a tendency for linked transposition that can be exploited in regional mutagenesis experiments (Hake et al. 1989; Athma et al. 1992; Moreno et al. 1992; Weil et al. 1992; Alleman and Kermicle 1993; Colasanti et al. 1998) and to obtain additional gene sequence from closely linked somatic transpositions (Singh et al. 2003).
Although Ac and Ds share these common features a major difference is that Ac-induced mutations are genetically unstable. The use of Ds in a two-component tagging approach would allow for the generation of stable or mutable alleles. As outlined above, this can be useful in a range of studies from crop improvement to clonal analysis. Here, we have adapted the Ac-casting method to amplify sequences flanking Ds insertions following somatic transposition. We have also shown that it is possible to select for novel Ds derivatives from multiple Ac elements throughout the genome. Each of the Ds elements identified carries a unique molecular signature consisting of either filler DNA, a novel breakpoint, or a combination of the two. These unique molecular tags can be readily exploited in PCR-based techniques to selectively amplify DNA flanking each unique Ds element. For instance, we are currently amplifying germinal transpositions of the Ds resident at the r1-sc:m3 locus, by exploiting the unique breakpoint in the Ds6-like element resident as a molecular signature in IPCR (http://www.plantgdb.org/prj/AcDsTagging/).
Two component systems have been used for gene characterization extensively in Arabidopsis, tomato and rice (for example, Bancroft et al. 1993; Jones et al. 1994; James et al. 1995; Sundaresan et al. 1995; Chin et al. 1999; Meissner et al. 1999). However, in maize only two genes, Indeterminate1 and Knotted1, have been cloned using a two-component directed tagging approach with Ds (Hake et al. 1989; Colasanti et al. 1998). One of the primary factors limiting Ds tagging in maize is the lack of molecularly mapped Ds insertions in the genome. By definition, a Ds element does not encode an active transposase, and this has made it difficult to develop genetic screens to mobilize these elements. However, several groups have successfully distributed Ac elements throughout the genome (Auger and Sheridan 1999; Cowperthwaite et al. 2002; Kolkman et al. 2005). Currently, 171 Ac elements distributed throughout the maize genome are available from the Maize Genetics Cooperative Stock Center (http://www.maizegdb.org/cgi-bin/stockcatalog.cgi#tool). Thus, one approach to distributing Ds insertions throughout the maize genome is to select for Ds derivatives from mapped Ac elements using the methods described here.
To demonstrate the utility of a two-component tagging approach, we identified five Ds insertions at the ps1 locus of maize. The ps1-m8∷Ac allele conditions a weak mutant phenotype that accumulates lycopene in the embryo due to an Ac insertion in the 5′-UTR of the lycopene β-cyclase gene (Bai et al. 2007). Ds insertions at this locus provide a viable stable mutant phenotype that can now be used to move the increased lycopene trait into maize breeding lines. Additionally, these Ds insertions can be used as donor elements to create numerous stable Ds insertions throughout this gene for further structure/function studies. One derivative, ps1-m8∷Ds(D2), was identified that contained a single intact end of the Ac element, known as a fractured Ac. These elements have been shown to induce chromosome breakage and more complex rearrangements when linked in cis with a functional Ac element (Dooner and Belachew 1991; Zhang and Peterson 2004). Thus, it should now be possible to generate deletion derivatives and a number of stable ps1 derivatives using this series of Ds elements.
Frequency of Ds element formation:
In this study we identified 15 Ds derivatives from Ac insertions at eight loci. The frequency of Ds formation across these eight loci was ∼0.025%. This estimate is similar to frequency of Ds formation calculated by Dooner and colleagues at bz1-m2∷Ac (2/3867 × 100 = 0.05% (Yan et al. 1999). However, McClintock identified 13 putative Ds derivatives from the wx-m9∷Ac allele in screens of 4737 gametes (McClintock 1963). She conducted further genetic tests on only two of these derivatives and confirmed that both were Ds derivatives. Thus, the frequency of Ds formation from wx-m9∷Ac may be as high as 0.27% and is minimally 0.04%. Interestingly, we identified a single Ds derivative in screens of only 895 progeny from bti00228∷Ac, suggesting that the frequency of Ds formation can vary as a function of the Ac insertion site. However, in the absence of additional Ds derivatives from bti00228∷Ac or an accurate frequency of Ds formation from wx-m9∷Ac, we can only speculate on the significance of these findings. Studies of Ac excision in maize suggest that immediately following Ac excision, a double-strand break is formed resulting in hairpin formation of flanking DNA (Scott et al. 1996; Bai et al. 2007). These hairpins are likely resolved through endonucleolytic attack resulting in predominant 7- and 8-bp excision alleles (Bai et al. 2007). In rare instances, hairpin formation does not occur and instead the excision site is repaired through DNA synthesis using the sister chromatid as a template (Rubin and Levy 1997; Yan et al. 1999). If repair does not execute faithfully, a derivative Ds element may be produced. It is possible that local chromatin organization or epigenetic modifications can influence the DNA repair process following Ac excision and therefore influence the frequency of Ds element formation.
Model for Ds formation:
All 15 Ds elements sequenced in this study have a unique deletion junction with the insertion of filler DNA occurring in about half of them (7/15). The filler DNA originates from sequence near the deletion and in one instance comes from genomic sequences flanking the Ds insertion. In previous reports of Ds formation, many Ds derivatives contain filler DNA at the deletion junction that originated from sequence nearby, within, and outside Ac (Rubin and Levy 1997; Yan et al. 1999). The occurrence of filler DNA and direct repeats in the template element flanking deleted sequences has also been observed at the deletion breakpoints of nonautonomous transposons derived from P elements in Drosophila (O'hare and Rubin 1983; Takasu-Ishikawa et al. 1992) and Mutator in maize (Hsia and Schnable 1996), suggesting that the insertion of filler DNA is common in the repair of transposon-mediated excision events. As mentioned above, filler DNA results from mispairing during DNA synthesis following Ac excision (for reviews see Gorbunova and Levy 1999; Puchta 2005). The common feature of filler DNA within Ds elements is that the choice of template is dependent on its position relative to the deletion breakpoint.
Interestingly, all of the newly derived Ds derivatives identified by Yan et al. (1999), contained short direct repeat sequences in the template Ac element at the deletion breakpoints of the Ds, suggesting repeats were involved in the mechanism of Ds formation. In studies of native Ds elements in the maize genome, the majority also carried simple deletions that appear to originate at short direct repeats within Ac. However, several Ds insertions were also defined with deletion breakpoints that do not appear to have arisen from simple slip mispairing during repair synthesis (Pohlman et al. 1984; Dooner 1986; Rubin and Levy 1997). Thus, it was unclear if the lack of repeat sequences in the Ac element at the Ds deletion junction was due to secondary mutations or simply the outcome of another repair process.
In contrast to these previous reports, several deletion junctions or filler DNA sequences in this study did not include a single copy sequence that is repeated twice in the progenitor element at the deletion junction (minimally 5/15). Moreover, several of these Ds derivatives contain filler DNA originating from multiple locations within Ac indicating a nonsequential positioning of filler during repair synthesis. For instance, ps1-m8∷Ds(D3) contains five blocks of filler DNA derived from upstream, downstream, and within the deleted region of Ac. This suggests that slip mispairing may be occurring on both upper and lower template strands during DNA synthesis and that slip mispairing may occur at least five times during the synthesis of one Ds element. Alternatively, these complex insertions of filler DNA could be created through multiple, independent rounds of excision and repair. However, since the Ds elements in this study formed in a single generation, it is highly unlikely that multiple excision events occurred.
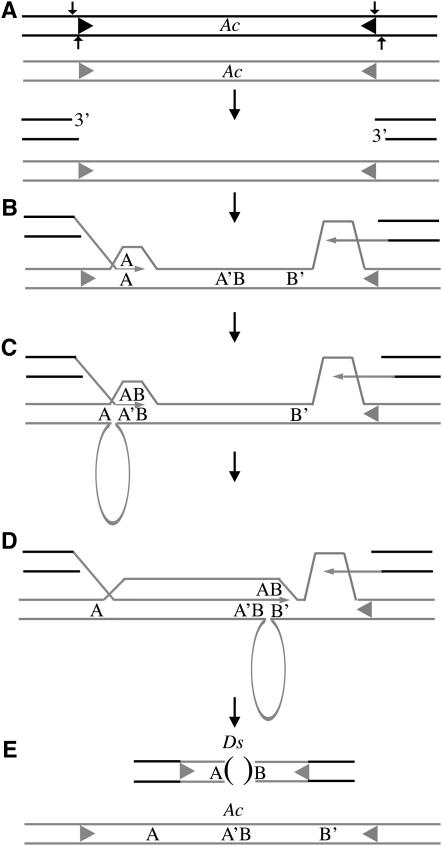
The most recent model for Ds formation attributes both deletion formation and insertion of filler DNA to slip mispairing at direct repeat sequences within the template Ac (Yan et al. 1999). However, this model cannot account for the formation of 5/15 Ds elements identified in this study. Figure 5 illustrates a model of Ds formation that demonstrates how slip mispairing may occur without direct repeats at the deletion junction in the template DNA. Previous studies have established that Ds formation is dependent upon the excision of Ac (Rubin and Levy 1997). Thus, the process of Ds formation likely begins with Ac excision, initiating a DSB (Figure 5A; reviewed in Kunze and Weil 2002). Typically, this double-strand break is repaired through a hairpin intermediate resulting in Ac footprint formation (Weil and Kunze 2000; Bai et al. 2007). However, in rare instances, the DSB is resolved through 3′ strand invasion that initiates a SDSA repair pathway with as little as 1–6 bp of homology required to initiate synthesis (Rubin and Levy 1997; Salomon and Puchta 1998; Gorbunova and Levy 1999; Puchta 2005).
Figure 5.—
Model for Ds formation. (A) A DSB is created at the donor locus through Ac excision. Arrows indicate a staggered cut resulting in a 1-bp 5′ overhang (review in Kunze and Weil 2002). The sister chromatid is shown as shaded. (B) A SDSA mechanism is initiated through a 3′-strand invasion of the sister chromatid. DNA synthesis begins from both excision sites using the sister chromatid as a template for repair. Letters (A, A′ and B, B′) represent 1–5 bp of sequences that occur multiple times within Ac. (C) Slippage occurs at sequence motif A with reannealing at A′ and a deleted segment of the Ac template (looped-out region) during DNA synthesis. Filler DNA sequence B is copied into the new Ds element now adjacent to sequence A rather than A′ as occurs in the template. (D) A second slip mispairing occurs at sequences B with reannealing at B′ resulting in another deletion of template DNA (looped-out region) in the new Ds element. (E) Newly synthesized DNA is ligated back into the excision site via NHEJ. The Ds product carries a unique AB junction not present in the donor Ac.
We envision DNA repair synthesis initiating from both ends of the excised element using the sister chromatid as a template (Figure 5B). Typically, slip mispairing takes place at short stretches of sequences that occur multiple times within the template Ac (Rubin and Levy 1997; Yan et al. 1999) and it has been suggested that 1–5 bp of homology is sufficient to invoke slippage during DNA synthesis (Yan et al. 1999). For simplicity these repeat sequences are represented by letters A, A′, B, and B′ in Figure 5, B–E. Following DNA synthesis of sequence A (e.g., 3-bp sequence), slip mispairing occurs followed by reannealing at A′, resulting in a deletion between the repeat sequence A and A′ in the Ac template. This single slip- mispairing event would result in the generation of the type I Ds elements shown in Figure 4 and those previously characterized by Yan et al. (1999). However, we also observed sequential slippage events, as illustrated in Figure 5D. Here, the short filler DNA sequence B (e.g., 3 bp) is copied into the new Ds element adjacent to sequence A (Figure 5C). Immediately following synthesis of sequence B, a second slip mispairing occurs, resulting in reannealing at B′ and another deletion of template DNA in the new Ds elements (Figure 5D) generating a novel junction AB. DNA synthesis is concluded when the template disassociates from the newly synthesized DNA and the molecule is ligated to the excision site via nonhomologous end joining (NHEJ) (Figure 5E). As shown in Figure 5E, the end result is a Ds element with a deletion flanked by sequences A and B. However, in the template strand these sequences are not contiguous; they appear as A, A′B, and B′. Thus, by invoking a series of tandem slip-mispairing events, this model can accommodate the formation of both type I and type II Ds elements observed in our study (Figure 4).
In summary, our data agree with previous models suggesting that error-prone DNA repair initiated through the SDSA pathway gives rise to Ds derivatives following Ac excision (Rubin and Levy 1997; Yan et al. 1999). However, our data suggest slip mispairing can occur numerous times during DNA repair synthesis, resulting in complex mosaics of filler DNA that are derived from Ac sequences located both in front of and behind the replication fork as observed in ps1-m8∷Ds(D1) and ps1-m8∷Ds(D3). Previous models of Ds formation do not invoke multiple rounds of slippage nor do they suggest a reordering of template DNA copied into the deletion junction as was observed here. Although we could invoke NHEJ or abortive gap repair to explain some type II Ds formation, we prefer the more parsimonious model described above that can account for the formation of all state II Ds elements.
Acknowledgments
We acknowledge Marika Olson for conducting molecular screens to identify the bti00191∷Ds element. We thank Prasit Deewatthanawong for the 96-well DNA extraction protocol. We also thank Judy Kolkman, Tracy Blasioli, and Phyllis Farmer for providing seed for screening of derivative alleles. We are grateful to Amanda Romag, Andrew Florino, and Lauren Pitt for assistance with tissue collection and genetic analysis. We also thank Erik Vollbrecht and Kazuhiro Kikuchi for critical readings of the manuscript. This work was supported by funding from the National Science Foundation to T.P.B. (DBI-0501713).
References
- Alleman, M., and J. L. Kermicle, 1993. Somatic variegation and germinal mutability reflect the position of transposable element Dissociation within the maize R gene. Genetics 135: 189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athma, P., E. Grotewold and T. Peterson, 1992. Insertional mutagenesis of the maize P gene by intragenic transposition of Ac. Genetics 131: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger, D. L., and W. Sheridan, 1999. Maize stocks modified to enhance the recovery of Ac-induced mutations. J. Hered. 90: 453–458. [Google Scholar]
- Bai, L., M. Singh, L. Pitt, M. Sweeney and T. P. Brutnell, 2007. Generating novel allelic variation through Activator insertional mutagenesis in maize. Genetics 175: 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, I., J. D. Jones and C. Dean, 1993. Heterologous transposon tagging of the DRL1 locus in Arabidopsis. Plant Cell 5: 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens, U., N. Fedoroff, A. Laird, M. Muller-Neumann, P. Starlinger et al., 1984. Cloning of the Zea mays controlling element Ac from the wx-m7 allele. Mol. Gen. Genet. 194: 346–347. [Google Scholar]
- Brutnell, T. P., 2002. Transposon tagging in maize. Funct. Integr. Genomics 2: 4–12. [DOI] [PubMed] [Google Scholar]
- Brutnell, T. P., and L. J. Conrad, 2003. Transposon tagging using Activator (Ac) in maize. Methods Mol. Biol. 236: 157–176. [DOI] [PubMed] [Google Scholar]
- Brutnell, T. P., and S. L. Dellaporta, 1994. Somatic inactivation and reactivation of Ac associated with changes in cytosine methylation and transposase expression. Genetics 138: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell, T. P., B. P. May and S. L. Dellaporta, 1997. The Ac-st2 element of maize exhibits a positive dosage effect and epigenetic regulation. Genetics 147: 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock, P., and P. J. Hooykaas, 1996. Integration of Agrobacterium tumefaciens T-DNA in the Saccharomyces cerevisiae genome by illegitimate recombination. Proc. Natl. Acad. Sci. USA 93: 15272–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., and S. L. Dellaporta, 1994. Urea-based plant DNA miniprep, pp. 526–527 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Chin, H. G., M. S. Choe, S. H. Lee, S. H. Park, J. C. Koo et al., 1999. Molecular analysis of rice plants harboring an Ac/Ds transposable element-mediated gene trapping system. Plant J. 19: 615–623. [DOI] [PubMed] [Google Scholar]
- Chomet, P. S., S. Wessler and S. L. Dellaporta, 1987. Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 6: 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti, J., Z. Yuan and V. Sundaresan, 1998. The Indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603. [DOI] [PubMed] [Google Scholar]
- Conrad, L. J., and T. P. Brutnell, 2005. Ac-immobilized, a stable source of Activator transposase that mediates sporophytic and gametophytic excision of Dissociation elements in maize. Genetics 171: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland, G., B. Baker, J. Schell and P. Starlinger, 1988. Characterization of the maize transposable element Ac by internal deletions. EMBO J. 7: 3653–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupland, G., C. Plum, S. Chatterjee, A. Post and P. Starlinger, 1989. Sequences near the termini are required for transposition of the maize transposon Ac in transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 86: 9385–9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite, M., W. Park, Z. Xu, X. Yan, S. C. Maurais et al., 2002. Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R. K., and M. Freeling, 1990. Clonal analysis of the cell lineages in the male flower of maize. Dev. Biol. 142: 233–245. [DOI] [PubMed] [Google Scholar]
- Dooner, H. K., 1986. Genetic fine structure of the bronze locus in maize. Genetics 113: 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., and A. Belachew, 1989. Transposition pattern of the maize element Ac from the Bz-m2(Ac) allele. Genetics 122: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., and A. Belachew, 1991. Chromosome breakage by pairs of closely linked transposable elements of the Ac-Ds family in maize. Genetics 129: 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring, H. P., M. Freeling, S. Hake, M. A. Johns, R. Kunze et al., 1984. A Ds mutation of the Adh1 gene in Zea mays L. Mol. Gen. Genet. 193: 199–204. [Google Scholar]
- Doring, H. P., B. Nelsen-Salz, R. Garber and E. Tillmann, 1989. Double Ds elements are involved in specific chromosome breakage. Mol. Gen. Genet. 219: 299–305. [DOI] [PubMed] [Google Scholar]
- Doring, H. P., I. Pahl and M. Durany, 1990. Chromosomal rearrangements caused by the aberrant transposition of double Ds elements are formed by Ds and adjacent non-Ds sequences. Mol. Gen. Genet. 224: 40–48. [DOI] [PubMed] [Google Scholar]
- Emerson, R., 1917. Genetical studies of variegated pericarp in maize. Genetics 2: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff, N., S. Wessler and M. Shure, 1983. Isolation of the transposable maize controlling elements Ac and Ds. Cell 35: 235–242. [DOI] [PubMed] [Google Scholar]
- Fusswinkel, H., S. Schein, U. Courage, P. Starlinger and R. Kunze, 1991. Detection and abundance of mRNA and protein encoded by transposable element Activator (Ac) in maize. Mol. Gen. Genet. 225: 186–192. [DOI] [PubMed] [Google Scholar]
- Giroux, M. J., J. Shaw, G. Barry, B. G. Cobb, T. Greene et al., 1996. A single mutation that increases maize seed weight. Proc. Natl. Acad. Sci. USA 93: 5824–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova, V. V., and A. A. Levy, 1999. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4: 263–269. [DOI] [PubMed] [Google Scholar]
- Greenblatt, I. M., 1984. A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, Modulator, in maize. Genetics 108: 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake, S., E. Vollbrecht and M. Freeling, 1989. Cloning Knotted, the dominant morphological mutant in maize using Ds2 as a transposon tag. EMBO J. 8: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia, A. P., and P. S. Schnable, 1996. DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics 142: 603–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, Jr., D. W., E. Lim, J. Keller, I. Plooy, E. Ralston et al., 1995. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon Activator. Plant Cell 7: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D. A., C. M. Thomas, K. E. Hammond-Kosack, P. J. Balint-Kurti and J. D. Jones, 1994. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266: 789–793. [DOI] [PubMed] [Google Scholar]
- Kolkman, J., L. J. Conrad, P. R. Farmer, K. Hardeman, K. R. Ahern et al., 2005. Distribution of Activator (Ac) throughout the maize genome for use in regional mutagenesis. Genetics 169: 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., H. Saedler and W.-E. Lönnig, 1997. Plant transposable elements, pp. 332–469 in Advances in Botanical Research, edited by J. A. Callow. Academic Press, London.
- Kunze, R., P. Starlinger and D. Schwartz, 1988. DNA methylation of the maize transposable element Ac interferes with its transcription. Mol. Gen. Genet. 214: 325–327. [Google Scholar]
- Kunze, R., U. Stochaj, J. Laufs and P. Starlinger, 1987. Transcription of the transposable element Activator (Ac) of Zea mays L. EMBO J. 6: 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., and C. F. Weil, 2002. The hAT and CACTA superfamily of plant transposons, pp. 565–610 in Mobile DNA, edited by N. L. Craig. ASM, Washington, DC.
- McClintock, B., 1946. Maize Genetics. Carnegie Inst. Wash. Year Book 45: 176–186. [PubMed] [Google Scholar]
- McClintock, B., 1949. Mutable loci in maize. Carnegie Inst. Wash. Year Book 48: 142–154. [PubMed] [Google Scholar]
- McClintock, B., 1951. Chromosome organization and gene expression. Cold Spring Harbor Symp. Quant. Biol. 16: 13–47. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1955. Controlled mutation in maize. Carnegie Inst. Wash. Year Book 54: 245–255. [PubMed] [Google Scholar]
- McClintock, B., 1956. Mutation in maize. Carnegie Inst. Wash. Year Book 55: 323–332. [PubMed] [Google Scholar]
- McClintock, B., 1962. Topographical relations between elements of control systems in maize. Carnegie Inst. Wash. Year Book 61: 448–461. [Google Scholar]
- McClintock, B., 1963. Further studies of gene-control systems in maize. Carnegie Inst. Wash. Year Book 62: 486–493. [Google Scholar]
- Meissner, R. C., H. Jin, E. Cominelli, M. Denekamp, A. Fuertes et al., 1999. Function search in a large transcription factor gene family in Arabidopsis: assessing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell 11: 1827–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, M. A., J. Chen, I. Greenblatt and S. L. Dellaporta, 1992. Reconstitutional mutagenesis of the maize P gene by short-range Ac transpositions. Genetics 131: 939–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Neumann, M., J. I. Yoder and P. Starlinger, 1984. The DNA sequence of the transposable element Ac of Zea mays L. Mol. Gen. Genet. 198: 9–24. [Google Scholar]
- O'Hare, K., and G. Rubin, 1983. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell 34: 25–35. [DOI] [PubMed] [Google Scholar]
- Pohlman, R. F., N. V. Fedoroff and J. Messing, 1984. The nucleotide sequence of the maize controlling element Activator. Cell 37: 635–643. [DOI] [PubMed] [Google Scholar]
- Puchta, H., 2005. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56: 1–14. [DOI] [PubMed] [Google Scholar]
- Ralston, E., J. English and H. K. Dooner, 1989. Chromosome-breaking structure in maize involving a fractured Ac element. Proc. Natl. Acad. Sci. USA 86: 9451–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, D. B., X. B. Chang and J. H. Wilson, 1989. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol. Cell. Biol. 9: 3049–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, E., and A. A. Levy, 1997. Abortive gap repair: underlying mechanism for Ds element formation. Mol. Cell. Biol. 17: 6294–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsard-Chanet, A., and O. Begel, 1990. Insertion of an LrDNA gene fragment and of filler DNA at a mitochondrial exon-intron junction in Podospora. Nucleic Acids Res. 18: 779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon, S., and H. Puchta, 1998. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 17: 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser, L., A. Roussis, J. Stiller and J. Stougaard, 1999. A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Schultes, N. P., T. P. Brutnell, A. Allen, S. L. Dellaporta, T. Nelson et al., 1996. Leaf permease1 gene of maize is required for chloroplast development. Plant Cell 8: 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D., and E. Dennis, 1986. Transposase activity of the Ac controlling element in maize is regulated by its degree of methylation. Mol. Gen. Genet. 205: 476–482. [Google Scholar]
- Scott, L., D. LaFoe and C. F. Weil, 1996. Adjacent sequences influence DNA repair accompanying transposon excision in maize. Genetics 142: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles, A. M., 2005. Maize community resources for forward and reverse genetics. Maydica 50: 405–414. [Google Scholar]
- Singh, M., P. E. Lewis, K. Hardeman, L. Bai, J. K. Rose et al., 2003. Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell 15: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V., P. Springer, T. Volpe, S. Haward, J. D. Jones et al., 1995. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- Sutton, W., W. Gerlach, D. Schwartz and W. Peacock, 1984. Molecular analysis of Ds controlling element mutations at the Adh1 locus of maize. Science 223: 1265–1268. [DOI] [PubMed] [Google Scholar]
- Takasu-Ishikawa, E., M. Yoshihara and Y. Hotta, 1992. Extra sequences found at P element excision sites in Drosophila melanogaster. Mol. Gen. Genet. 232: 17–23. [DOI] [PubMed] [Google Scholar]
- Van Schaik, N. W., and R. A. Brink, 1959. Transposition of Modulator, a component of the variegated pericarp allele in maize. Genetics 44: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona, M., and S. R. Wessler, 1990. Implications for the cis-requirements for Ds transposition based on the sequence of the wxB4 Ds element. Mol. Gen. Genet. 220: 414–418. [DOI] [PubMed] [Google Scholar]
- Weck, E., U. Courage, H. P. Doring, N. Fedoroff and P. Starlinger, 1984. Analysis of sh-m6233, a mutation induced by the transposable element Ds in the sucrose synthase gene of Zea mays. EMBO J. 3: 1713–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil, C. F., and R. Kunze, 2000. Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat. Genet. 26: 187–190. [DOI] [PubMed] [Google Scholar]
- Weil, C. F., S. Marillonnet, B. Burr and S. R. Wessler, 1992. Changes in state of the Wx-M5 allele of maize are due to intragenic transposition of Ds. Genetics 130: 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil, C. F., and S. R. Wessler, 1993. Molecular evidence that chromosome breakage by Ds elements is caused by aberrant transposition. Plant Cell 5: 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S., A. Tarpley, M. Purugganan, M. Spell and R. Okagaki, 1990. Filler DNA is associated with spontaneous deletions in maize. Proc. Natl. Acad. Sci. USA 87: 8731–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S. R., G. Baran, M. Varagona and S. L. Dellaporta, 1986. Excision of Ds produces waxy proteins with a range of enzymatic activities. EMBO J. 5: 2427–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, S., T. Takano-Shimizu, K. Kitamura, T. Mikami and Y. Kishima, 1999. Resistance to gap repair of the transposon Tam3 in Antirrhinum majus: a role of the end regions. Genetics 153: 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X., I. M. Martinez-Ferez, S. Kavchok and H. K. Dooner, 1999. Origination of Ds elements from Ac elements in maize: evidence for rare repair synthesis at the site of Ac excision. Genetics 152: 1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and T. Peterson, 2004. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics 167: 1929–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]