Abstract
Molecular changes that underlie evolutionary changes in behavior and physiology are not well understood. Dauer formation in Caenorhabditis elegans is a temperature-sensitive process controlled through a network of signaling pathways associated with sensory neurons and is potentially an excellent system in which to investigate molecular changes in neuronal function during evolution. To begin to investigate the evolution of dauer formation in the genus Caenorhabditis at the molecular level, we isolated dauer-formation mutations in C. briggsae, a species closely related to the model organism C. elegans. We identified mutations in orthologs of C. elegans genes daf-2 (insulin receptor), daf-3 (Smad), and daf-4 (TGF-β type 2 receptor), as well as genes required for formation of sensory cilia. Phenotypic analyses revealed that functions of these genes are conserved between C. elegans and C. briggsae. Analysis of C. briggsae mutations also revealed a significant difference between the two species in their responses to high temperatures (>26°). C. elegans is strongly induced to form dauers at temperatures above 26°, near the upper limit for growth of C. elegans. In contrast, C. briggsae, which is capable of growth at higher temperatures than C. elegans, lacks this response.
CAENORHABDITIS briggsae is a nematode closely related to the model organism C. elegans (reviewed in Gupta et al. 2007). A range of molecular, genetic, and genomic resources makes this species an ideal target of comparative studies with C. elegans. The genome of C. briggsae has been sequenced and is annotated in detail (Stein et al. 2003). Since C. briggsae propagates as self-fertilizing hermaphrodites like C. elegans, genetic analysis of C. briggsae is facile, and a number of labs have produced genetic tools [e.g., morphological markers or single nucleotide polymorphism markers (Hillier et al. 2007)] that are useful in genetic mapping and strain construction (Gupta et al. 2007). Molecular manipulations including transformation (Kennedy et al. 1993; Kirouac and Sternberg 2003) and RNAi (Rudel and Kimble 2001; Nayak et al. 2005) are possible in C. briggsae. Like C. elegans, C. briggsae has nearly invariant cell lineages that produce identical sets of cells in every animal. The complement of cells is also conserved between C. elegans and C. briggsae, providing an opportunity to compare functions of individual cells (Delattre and Felix 2001).
Despite their similarities, C. briggsae and C. elegans are distinct species with overlapping geographic distributions and probably occupy different ecological niches (Cutter et al. 2006). One known difference of possible ecological significance is in their temperature responses. All isolates of C. elegans grow at temperatures from ∼10° to 27° (M. Ailion and J. H. Thomas, unpublished data). In contrast, C. briggsae can be cultured at temperatures over 29°. This difference in temperature preferences suggested that dauer formation response might also differ between the two species. As with C. elegans (Cassada and Russell 1975), C. briggsae responds to unfavorable environmental conditions (especially high population density, absence of food bacteria, and high temperature; Golden and Riddle 1984b) by entering the dauer stage, which is a developmentally arrested third larval stage (Fodor et al. 1983). This response is mediated in part by dauer pheromones, secreted molecules that induce dauer formation in C. elegans and C. briggsae (Golden and Riddle 1982; Jeong et al. 2005; Butcher et al. 2007). Dauers exhibit specific morphological, physiological, and behavioral characteristics that enable them to survive harsh conditions and may aid in dispersal (Cassada and Russell 1975). When a favorable environment is encountered, dauers reenter the normal life cycle and go on to become normal adults. In C. elegans, dauer formation in the 15° to 25° range shows broad temperature sensitivity with higher temperatures promoting entry into the dauer stage (Golden and Riddle 1984a,b). Furthermore, at temperatures near the upper limit of growth and reproduction (>26°, typically assayed at 27°), there is exceptionally strong induction of dauer formation (Ailion and Thomas 2000). Several lines of evidence indicate that this high-temperature response is qualitatively distinct from temperature sensitivity in the 15° to 25° range. For example, some genes that promote dauer formation at lower temperatures inhibit dauer formation at higher temperatures (Ailion and Thomas 2000). The observation that C. briggsae is capable of growth at higher temperatures (>29°) raised an interesting question: Does C. briggsae exhibit a similar response, and if it does, at what temperature?
To begin to investigate evolutionary changes in mechanisms that regulate dauer formation, we isolated and characterized mutations in C. briggsae that affect dauer formation. Analogous mutations in C. elegans have been studied for many years (Figure 1) (Riddle et al. 1981; Thomas 1993). Together, these mutations enable us to compare the functions of dauer-regulating genes in the two species. From several screens we identified mutants that fail to form dauers normally when exposed to dauer-inducing conditions (dauer-formation defective or Daf-d mutants) and mutants that form dauers even in dauer noninducing conditions (dauer-formation constitutive or Daf-c mutants). Phenotypic analyses and mapping suggested molecular identities for some mutations, and sequencing confirmed three to be mutations in orthologs of daf-2 (insulin receptor), daf-3 (Smad), and daf-4 (TGF-β type 2 receptor). Phenotypes and genetic interactions demonstrate that functions of these genes are conserved between C. elegans and C. briggsae. Furthermore, using these mutations, we tested high-temperature dauer-formation responses in C. briggsae. Although dauer formation in C. briggsae is broadly temperature sensitive, we found no evidence of dauer hyperinduction at high temperatures in C. briggsae.
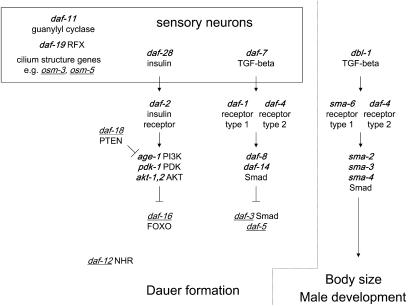
Figure 1.—
Genetic pathways regulating dauer formation in C. elegans. This figure shows a subset of genes that mutate to affect dauer formation. Genes that cause constitutive dauer formation when mutated are shown in boldface and genes that cause defective dauer formation are underlined. Genes that function in neurons are shown in the box at the top. Genes in each pathway (TGF-β, insulin) usually demonstrate clear epistasis relationships. Interactions between mutations that affect different pathways are complex. The paralogous pathway regulating body size and male tail development is shown to the right.
MATERIALS AND METHODS
Genetics and nomenclature:
We used the C. briggsae isolate AF16 (available from Caenorhabditis Genetics Center, University of Minnesota) as the wild-type strain. This strain is thought to exhibit wild-type dauer formation behavior (Fodor et al. 1983). All strains described in this article are in this background. Mutations used for mapping are LGI sma(sy5330), LGII cby-15(sy5148), Cb-unc-4(sy5341), LGIII cby-4(sy5016), LGIV mip-1(s1270), LGV unc(sa997) LGX cby-3(sy5039), rot-1(sy5001), and unc(sa988). cby-4 and mip-1 are thought to be orthologs to dpy-1 III and unc-22 IV respectively.
We use the guideline for nomenclature of non-C. elegans strains extended from the published guideline for C. elegans (Horvitz et al. 1979; Hodgkin 1995). Briefly, strain names and mutation names (allele numbers) follow the same format as C. elegans. There is no overlap with C. elegans strain and allele designations. Where orthology to a C. elegans gene is known, the gene name reflects the orthology, e.g., Cb-daf-4 is the C. briggsae ortholog of C. elegans daf-4. Different gene/phenotype names are used for C. briggsae genes whose orthology has not been determined. Corresponding names are (C. briggsae/C. elegans): Cby (chubby)/Dpy (dumpy), short and fat; Rot (rotator)/Rol (roller), spirally twisted animals; Mip (movement impaired)/Unc (uncoordinated), abnormal movement. Orthology and sequence information were obtained from WormBase (data freeze WS140). Cb-daf-2, Cb-daf-3, and Cb-daf-4 are CBG15732, CBG08108, and CBG08963.
Isolation and characterization of C. briggsae Daf-d mutations:
Ethylmethanesulfonate (EMS) mutagenesis was carried out as described for C. elegans, except in some cases animals were incubated in EMS solution for three hours instead of four (Brenner 1974). Mutagenized worms were allowed to self for two generations. F2 animals were picked one per plate. These plates were allowed to grow until starvation, at which time dauer formation was tested by flooding the plate with 1% SDS (sodium dodecyl sulfate) solution in water. All larvae except dauers are killed by 1% SDS in ∼15 min (Cassada and Russell 1975). Plates on which all animals were killed by SDS were identified as candidate Daf-d mutations. To allow recovery of such Daf-d mutations, a replicate of each plate was made by transferring a small piece of agar with worms to a new plate (chunking) prior to the SDS test. On further retests, we found that some mutations caused a complete block of dauer formation, whereas others formed some dauers under starvation conditions. We focused our analysis primarily on mutations that completely blocked dauer formation.
To test for the morphology of dauers, animals were mounted for Nomarski observation (Sulston and Horvitz 1977) and scored for dauer alae and modified pharynx. To test for a dye-filling defect, animals were stained with the DiI solution as described (Prasad et al. 1998).
To isolate phenotypic suppressors of the Daf-c mutation Cb-daf-4(sa973), we mutagenized with EMS a strain carrying Cb-daf-4(sa973). The F1 population was allowed to self at the permissive temperature (20°), and then the plates were shifted to 28°. Non-dauer F2's were picked, selfed to establish a line, and retested for dauer formation at 28°. From a screen of 4200 mutagenized genomes, we recovered eight independent mutations, of which one (sy5321) was analyzed in detail. sy5321 suppresses the Daf-c phenotype but not the Sma phenotype at the restrictive temperature for Cb-daf-4(sa973). Cb-daf-4(sa973); sy5321 animals were crossed to wild-type males, and progeny were allowed to self. Twenty-four randomly selected F2 animals were picked individually and allowed to self at 28°. Plates that had no dauers or Sma animals were allowed to grow until starvation and tested for the presence of dauers. Lines that segregated no Sma animals and did not form dauers were retained as an outcrossed sy5321 strain.
To map Daf-d mutations to the X chromosome, we crossed AF16 males to daf-d/daf-d hermaphrodites, and male progeny were crossed to an X-linked marker strain, most often PS9353 cby-3(sy5039) rot-1(sy5001) X. Cross-progeny hermaphrodites were allowed to self, and a number of phenotypically wild-type F2 progeny were picked individually to ∼20 plates. In the next generation, we selected plates that segregated no Cby Rots. These were allowed to starve, and examined for the presence of dauers. If the mutation was X-linked, very few or no dauers were observed. If the mutation was not X-linked, a majority of the plates segregated a large number of dauers.
The ability of daf-d mutations to suppress Cb-daf-4(sa973) was tested as follows. daf-4/+ males were crossed to daf-d/daf-d homozygotes. Progeny were picked to individual plates and allowed to self at 28°, where daf-4/+ cross-progeny segregated dauers. If the daf-d mutation suppressed the Daf-c phenotype of daf-4, we would observe Sma non-Daf-c animals segregating on the plate (daf-4/daf-4; daf-d/daf-d animals: 1/16 of the total population). Such animals were picked individually and allowed to self at 28° to confirm the presence of daf-4 and to confirm suppression. This test assumes that the daf-d mutation suppresses the Daf-c but not the Sma phenotype. No mutation in either species is known to suppress both phenotypes of daf-4.
Isolation and characterization of C. briggsae Daf-c mutations:
Screens for Daf-c mutants in the wild-type background were carried out as described (Riddle 1977). Screens for revertants of Cb-daf-3 were carried out as follows. A Cb-daf-3(sy5417) strain was EMS mutagenized and allowed to self at 25° for two or more generations, until the plates were starved. Animals were washed off from starved plates and were treated with 1% SDS solution for 15 min to kill non-dauers. The surviving population was washed again in water and transferred to fresh plates seeded with Escherichia coli. After recovery, these animals were allowed to self for multiple generations until starved and reselected using 1% SDS. Animals recovered from the second round of selection were picked on to fresh plates, and dauer formation was tested visually at different temperatures. Because the selection was carried out in bulk, and not every recovered animal was picked at the end, the number of genomes screened is unknown.
To map Cb-daf-2(sy5445), we allowed sy5445/cby-4 heterozygotes to self at 28°. Dauer progeny (sy5445 homozygotes) were picked and allowed to recover at 20°. Recovered dauers were selfed at 20°, and segregation of Cby (Dpy) animals was scored. None of seven sy5445 homozygotes segregated cby-4, indicating linkage to chromosome 3. To test whether Cb-daf-2(sy5445) extends lifespan, we picked mutant and wild-type animals in the L4 stage to plates seeded with OP50. Animals were transferred to fresh plates as necessary, for example because of overcrowding by progeny. Plates were checked every day for deaths, scored by the lack of movement. Animals that died because of unnatural causes, e.g., desiccation on the side of the plate, were not counted.
Sequencing of Cb-daf-2(sy5445), Cb-daf-3(sy5417), and Cb-daf-4(sa973):
The MH2 (C-terminal) domain of Cb-daf-3 was amplified from animals bearing the sy5417 mutation using primers CB39 and CB40 (Table 1). The amplified fragment was sequenced directly using the primer CB41. The mutation destroys an SfaNI restriction site present in the wild-type genomic sequence. Therefore to confirm the wild-type and mutant sequences, we amplified genomic sequences from sy5417 and wild-type animals using CB39 and CB40 and digested the fragments with SfaNI. The fragment from the wild type but not the mutant was digested by SfaNI at this site, confirming the change. The screen from which daf-3(sy5417) was isolated also produced two additional X-linked Daf-d mutant lines, to which were assigned allele numbers sy5418 and sy5419. When these strains were tested for the presence of the SfaNI site, they were found to lack the SfaNI site. sy5418 and sy5419 also failed to complement sy5417. Thus Cb-daf-3(sy5417), sy5418, and sy5419 are the same mutation. Since sy5417, sy5418, and sy5419 are derived from different mutagenized P0 animals in the screen, this probably indicates that this was a spontaneous mutation present in the parent population at a small frequency. Another X-linked Daf-d mutant, sy5315 does not have the mutation at the SfaNI site and complements sy5417.
TABLE 1.
Primers used in this study
| Primer | Sequence | Gene |
|---|---|---|
| CB39 | GCACCTACAAACATGACTAC | Cb-daf-3 |
| CB40 | CTGTTGAGATCGTATGTAAC | Cb-daf-3 |
| CB41 | CATCCATGAACTTGTATGCTC | Cb-daf-3 |
| CB8 | ACCGACTGCCGACCCTGTTG | Cb-daf-4 |
| CB9 | CCCTCTACCGTATATTTCGG | Cb-daf-4 |
| CB11 | TAAGGTAATCATGAACCTATC | Cb-daf-4 |
| CB15 | GAAAATCGAGAAAACGGGAAC | Cb-daf-4 |
| CB101 | TCGGTGACGGAGACTGGAAA AGTATGCCC | Cb-daf-2 |
| CB102 | GGGTGTTCGGGCGGATACGG TCTTTTGCC | Cb-daf-2 |
| CB103 | TTCGAGGCGATGCGTTTGGCC CATGCGCC | Cb-daf-2 |
| CB104 | TCATCCTTCTACCGCCCTTCC TCTCCTCC | Cb-daf-2 |
We sequenced the kinase domain of Cb-daf-4 from the mutant by amplifying the genomic sequence with primers CB8 and CB9 and sequencing the fragment directly with primers CB11 and CB15.
Although most daf-2 mutations in C. elegans reside in two large exons, the gene structure of Cb-daf-2 is different, and the homologous regions are spread out over a larger region of the genome (WormBase data freeze WS160). Therefore, we RT-PCR amplified these regions from mRNAs of mixed stage AF16 and sy5445 strains using primer pairs CB101/CB102 and CB103/CB104, and the PCR products were sequenced directly using the same primers. The sy5445 mutation was confirmed in the sequence of both strands. The same mutation is absent in our wild-type cDNA sequence and the published genomic sequence (WormBase data freeze WS160).
Rescue of Cb-daf-4 and generation of integrated lines:
To test rescue of the Cb-daf-4(sa973) mutant, we coinjected a Ce-daf-4 clone (Estevez et al. 1993) along with Ce-myo-2∷gfp as a coinjection marker (A. Fire, personal communication). Two stable transgenic lines were generated by following Ce-myo-2∷gfp. Transgenic animals were grown at restrictive temperatures (25° and 27°) to test for rescue. The integration of saEx[myo-2∷gfp, Ce-daf-4(+)] was done as described, using the rescue of the Cb-daf-4(sa973) Sma phenotype as the marker (Way and Chalfie 1989). Integrants were mapped using chromosomal markers sma(sy5330) I, cby-15 II, cby-4 III, and mip-1 IV.
RESULTS
Isolation and characterization of dauer formation defective mutants in C. briggsae:
We carried out a screen for dauer-formation defective mutations in the C. briggsae AF16 wild-type strain (Fodor et al. 1983). Since dauers (but not other stages) survive exposure to the 1% SDS solution (Cassada and Russell 1975; Riddle 1977), in an F2 clonal screen, we looked for lines that fail to survive in 1% SDS (see materials and methods). From a screen of ∼850 mutagenized genomes, we isolated 17 strong Daf-d mutations, of which 14 were kept (Table 2). In addition, the mutation sy5321 was isolated in a screen for suppressors of Cb-daf-4(sa973) (see materials and methods). Since sy5321 causes a Daf-d phenotype, it was analyzed in parallel. In C. elegans, daf-d mutants fall into several categories that exhibit distinct phenotypic characteristics. We carried out a set of tests to classify the newly identified C. briggsae mutants into analogous classes.
TABLE 2.
Daf-d mutants of C. briggsae
| Mutation | Gene | Class | Linkage | Dyf | Other |
|---|---|---|---|---|---|
| sy5311 | Cilium | III | Dyf(−) | ||
| sy5314 | Cilium | Dyf(−) | |||
| sy5420 | Cilium | Dyf(−) | |||
| sy5421 | Cilium | Dyf(−) | |||
| sy5313 | daf-13-like | Autosomal | |||
| sy5416 | daf-13-like | Autosomal | |||
| sy5429 | daf-13-like | ||||
| sy5417 | Cb-daf-3 | X | Dyf(+) | Suppresses Cb-daf-4(sa973) | |
| sy5315 | X | Dyf(+) | Makes partial dauers | ||
| sy5312 | Autosomal | Dyf(+) | |||
| sy5431 | Autosomal | Dyf(+) | |||
| sy5443 | Autosomal | Dyf(+) | |||
| sy5444 | Autosomal | Dyf(+) | |||
| sy5442 | Autosomal | ||||
| sa934 | Cilium | Autosomal | Dyf(−) | ||
| sy5321 | X | Dyf(+) | Cb-daf-4(sa973) suppressor |
sy5321 was isolated as a suppressor of Cb-daf-4(sa973). sa934 was isolated in a separate screen for Dyf mutants.
In C. elegans, the mutation daf-13(m66) causes animals to form morphologically normal dauers that fail to survive exposure to the 1% SDS solution (Riddle et al. 1981). We found that C. briggsae strains carrying sy5313, sy5416, or sy5429 exhibited the same phenotype (Figure 2). Isolation of multiple mutants of this class in C. briggsae is not surprising, since the screen for this type of mutants has not been saturated in C. elegans. These mutations likely affect specific physiological changes that make dauers more resistant to environmental stresses, but do not affect the mechanism that regulates dauer formation. These mutations were not analyzed further.
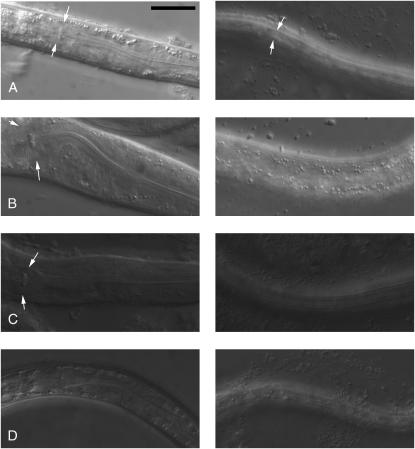
Figure 2.—
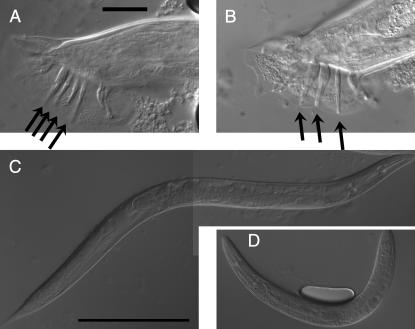
Partial dauer phenotypes of sy5313 and sy5315. Nomarski images of the pharynx (left) and dauer alae (or absence thereof, right) are shown. Arrows point to the terminal bulb of the pharynx (left) and dauer alae (right). All panels are shown at the same magnification. The scale bar is 20 μm. (A) Wild-type dauer. Pharynx is remodeled (narrower than in non-dauers; compare with panel B), and prominent dauer alae are present (visible as longitudinal stripes). (B) Wild-type non-dauer. Pharynx is wide and dauer alae are absent. (C) sy5315 partial dauer. Dauer alae are present but the pharynx is not fully remodeled. (D) sy5313 dauer. Both pharynx and alae are similar to wild-type dauers.
In C. elegans, mutation of any of a large group of genes (cilium structure genes) causes a defect in the structure of ciliated endings of sensory neurons (Perkins et al. 1986; Starich et al. 1995). Mutations affecting any of these genes cause a stereotypic set of phenotypes resulting from a general sensory defect. These include defects in dauer formation (Daf-d), osmotic avoidance (Osm), and chemotaxis (Che). Also, several sensory neurons in wild-type animals (both elegans and briggsae) fill with dyes (FITC, DiI, or DiO) when exposed to them in solution. In cilium structure mutants, the same cells are present, but fail to fill with dye (Dyf, dye-filling defective). In C. elegans, it has been reported that a large fraction of randomly isolated Daf-d mutations are of this class (Riddle 1977). We found mutations sy5311, sy5314, sy5420, and sy5421 were of this class on the basis of the dye-filling phenotype (Table 2) (see materials and methods). Another Daf-d mutation, sa934, was isolated in a pilot screen for Dyf mutants.
Many of the remaining mutations likely affect orthologs of daf-3 (Patterson et al. 1997), daf-5 (Da Graca et al. 2004), daf-12 (Antebi et al. 2000), daf-16 (Lin et al. 1997; Ogg et al. 1997), or daf-18 (Ogg and Ruvkun 1998; Gil et al. 1999; Mihaylova et al. 1999; Rouault et al. 1999); wherein the mutations cause the Daf-d phenotype (Riddle 1977; Riddle et al. 1981). Three types of experiments were carried out to help assign mutations to genes (Table 2). First, mutations were tested for linkage to the X chromosome. Chromosome-level synteny appears to be the rule for elegans–briggsae comparisons. [Genes on the same chromosome in C. elegans are usually found on the same chromosome in C. briggsae (Hillier et al. 2007).] Thus X-linked mutations could be alleles of daf-3 and daf-12 (X-linked in C. elegans; Riddle et al. 1981). Second, some of the mutations were tested for the ability to suppress Cb-daf-4(sa973). In C. elegans, mutations in daf-3, daf-5, and daf-12 completely suppress the Daf-c phenotype of daf-4 mutations (Riddle et al. 1981; Vowels and Thomas 1992; Thomas et al. 1993). We found that sy5417 suppressed Cb-daf-4(sa973). Third, strains bearing these mutations were tested for the ability to form partial dauers, animals that exhibit a subset of dauer-like characteristics. In C. elegans, all mutations in daf-16, daf-18, and some mutations in daf-12 cause animals to form partial dauers (Vowels and Thomas 1992). We found that the sy5315 bearing strain formed partial dauers in which dauer alae are present but the pharynx is not fully remodeled (Figure 2).
Although X-linked, sy5315 does not resemble typical daf-12 alleles, which do not form partial dauers. There is a unique partial dauer-forming daf-12 allele called m25, which is a mutation in the ligand binding domain of daf-12 (Antebi et al. 2000). We sequenced the ligand binding domain of Cb-daf-12 in a sy5315 bearing strain (not shown) and found no mutation.
Isolation and characterization of dauer formation constitutive mutations in C. briggsae:
In C. elegans, most Daf-c alleles, including many null alleles, are temperature sensitive for dauer entry because they do not completely eliminate the inherent temperature sensitivity of the pathway (Golden and Riddle 1984a; Malone and Thomas 1994). Such mutations also typically exhibit a temperature-sensitive defect in dauer recovery. Entry into the dauer stage is favored at high temperatures, while exit from the dauer stage is favored at lower temperatures. The original screen for Daf-c mutants in C. elegans utilized these characteristics (Riddle 1977), and we carried out an analogous screen in C. briggsae. We mutagenized wild-type animals, and dauers were isolated in the F2 generation at 25° or 28°, temperatures at which wild-type animals do not normally form dauers if not overcrowded and food is present. These dauers were allowed to recover at lower temperatures (20° or 15°) and selfed to establish a line. This screen requires that the mutant we isolate is capable of recovering from the dauer stage when downshifted to the lower temperature. Over 13,000 EMS mutagenized genomes were screened using this procedure and one strong Daf-c mutation, sa973, and several weaker mutations were isolated (Table 3; Figure 3). The frequency at which Daf-c mutants were recovered is much lower than the frequency of Daf-c mutations from a similar screen in C. elegans (Riddle 1977). Several factors might contribute to this difference. First, fewer genes in C. briggsae may mutate to cause the Daf-c phenotype. Second, a typical Daf-c mutation in C. briggsae may fail to recover when downshifted to 20° or 15°. Lastly, since different experimenters in different laboratories carried out these screens, there may be a difference in sensitivities to finding dauers from a large population.
TABLE 3.
Daf-c mutants of C. briggsae
| Mutation | Gene | Class | Linkage | Other |
|---|---|---|---|---|
| sa973 | Cb-daf-4 | III | ||
| sa974 | Weak Daf-c | |||
| sy5331 | Weak Daf-c | |||
| sy5367 | Weak Daf-c | |||
| sy5445 | Cb-daf-2 | III | Isolated as a daf-3 revertant |
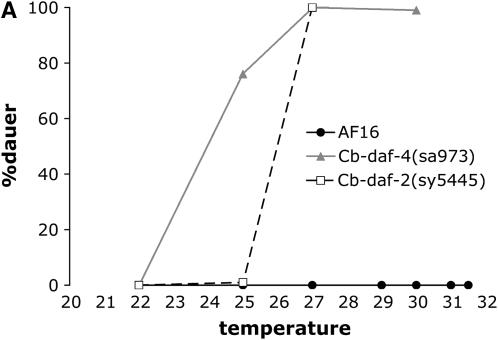
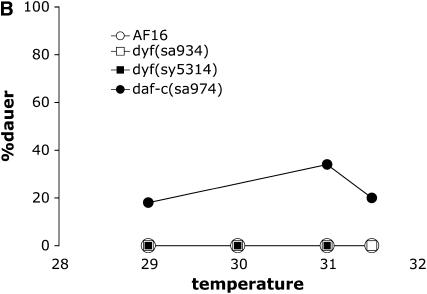
Figure 3.—
Dauer formation phenotypes of C. briggsae strains. (A) Dauer formation by strong Daf-c mutants. daf-4(sa973) strains are Sma (short and small) at 25° and above, but have normal body morphology at lower temperatures. (B) Dauer formation at high temperatures. Unlike in C. elegans, dyf mutants and weak daf-c mutants are not hyperinduced for dauer formation at these temperatures. Cb-daf-3(sy5417) and cby-15(sy5148) mutants also formed no dauers at 27° or 30° (not shown).
Because a screen for Daf-c mutations by a straightforward F2 screen failed to produce many mutants, we also carried out a screen for dauer-forming mutants in the Cb-daf-3 mutant background. Cb-daf-3(sy5417) mutants typically do not form any dauers, even at high population density and high temperature. If genetic interactions in C. briggsae are similar to those in C. elegans, we expect to recover mutations in genes daf-2 (Kimura et al. 1997), age-1 (Morris et al. 1996), pdk-1 (Paradis et al. 1999) (insulin pathway), daf-11 (guanylyl cyclase) (Birnby et al. 2000), and daf-19 (RFX transcription factor) (Swoboda et al. 2000). From these screens, we found one strong mutation, sy5445 (Table 3; Figure 3). Because the selection was carried out in bulk, the frequency of Daf-c mutations from this screen was not determined (see materials and methods).
sy5417 has a mutation in Cb-daf-3 (Smad):
The Daf-d mutation sy5417 is X-linked and suppresses the Daf-c phenotype of Cb-daf-4(sa973). Since in C. elegans, genes daf-3 and daf-12 map to the X-chromosome and mutations in these genes suppress daf-4(−), this suggested that sy5417 is a mutation in an ortholog of one of these genes (Riddle et al. 1981). daf-3 encodes a Smad protein, a cytoplasmic/nuclear transducer of TGF-β and related signals (Patterson et al. 1997). We sequenced the Cb-daf-3 gene from a strain carrying the sy5417 mutation (see materials and methods) and found that sy5417 has a missense mutation in the second conserved domain of Cb-daf-3 (Figure 4).
Figure 4.—
Molecular characterizations of Cb-daf-2, Cb-daf-3, and Cb-daf-4 mutations. A sequence alignment of each C. briggsae gene with the C. elegans ortholog and two or three other arbitrarily selected homologs is shown, and mutated residues are indicated below. The amino acid count is derived from the Wormbase WS150 gene model and may not reflect the actual gene structure.
sy5445 has a mutation in Cb-daf-2 (insulin receptor):
Genetic mapping placed the Daf-c mutation sy5445 on chromosome 3 (see materials and methods). C. elegans Daf-c genes daf-2, daf-4, and daf-7 map to chromosome 3. However, double mutants daf-4; daf-3 and daf-7; daf-3 are non-Daf-c (Riddle et al. 1981). Since sy5445 was isolated as a dauer constitutive mutation in the Cb-daf-3 mutant background, it was unlikely to be a mutation in Cb-daf-4 or Cb-daf-7. daf-2 encodes a homolog of an insulin receptor/receptor tyrosine kinase (Kimura et al. 1997). We sequenced the Cb-daf-2 cDNA from the sy5445 mutant (see materials and methods) and found a missense mutation in the kinase domain of Cb-daf-2 (Figure 4). The mutation affects a residue conserved in daf-2 and other insulin receptors, but not conserved in other receptor tyrosine kinases.
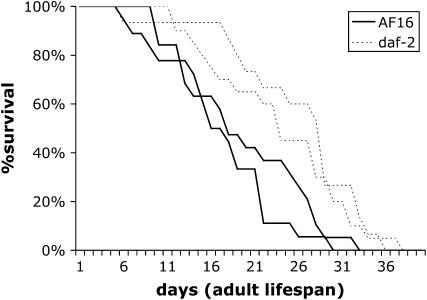
In C. elegans, mutations in daf-2 extend lifespan (Kenyon et al. 1993). To test whether the same was true in C. briggsae, we measured the survival of Cb-daf-2(sy5445) animals at 20°. The lifespan of wild-type AF16 animals (average ± standard deviation) was 19 ± 7.4 and 16 ± 6.5 in two trials. The lifespan of the Cb-daf-2 mutant was 25 ± 7.9 and 23 ± 7.8 in two trials done in parallel. Thus, as in C. elegans, a mutation in daf-2 increases the lifespan of C. briggsae (Figure 5).
Figure 5.—
Survival curves of AF16 wild type and Cb-daf-2(sy5445) at 20°. Two sets of assays were carried out for each strain. Numbers of animals are 19 and 18 for AF16 data sets and 15 and 20 for Cb-daf-2(sy5445) data sets. P-values were 0.0131 and 0.0119 (Mann-Whitney test).
sa973 is a mutation in Cb-daf-4 (TGF-β receptor type 2):
The Daf-c mutation sa973 exhibited a temperature-sensitive small-body-size phenotype (Sma). In addition, examination of males at restrictive temperatures revealed male tail defects, including crumpled spicules and ray defects (male abnormal; Mab) (Figure 6). In C. elegans, this combination of phenotypes (Sma, Mab, and Daf-c) is observed only in daf-4 mutants (Estevez et al. 1993; Savage et al. 1996). daf-4 encodes a type-2 TGF-β/BMP receptor (Estevez et al. 1993). We transformed the sa973 mutant strain with a Ce-daf-4 genomic clone (Estevez et al. 1993) and a transformation marker Ce-myo-2∷gfp (see materials and methods) and found that the transgene rescued the Sma phenotype. To determine the molecular nature of the defect, we sequenced the Cb-daf-4 gene from the sa973 mutant strain (see materials and methods) and found a mutation in the kinase domain (Figure 4). A typical C. elegans daf-4 mutation causes a temperature-sensitive Daf-c phenotype, whereas the Sma phenotype is not temperature sensitive (Estevez et al. 1993). Thus, the fact that the Cb-daf-4(sa973) Sma phenotype is temperature sensitive suggests that this missense mutation causes the CB-DAF-4 protein to be temperature sensitive.
Figure 6.—
Pleiotropic phenotypes of Cb-daf-4. (A) Lateral view of the wild-type male tail. Anterior is to the right. Arrows point to rays 3, 4, 5, and 6 (from right to left). The scale bar is 20 μm. (B) daf-4(sa973) male. The thicker ray in the middle is probably a result of ray 4/ray 5 fusion. (C) Wild-type hermaphrodite body shape. (D) The Sma phenotype of the daf-4(sa973) hermaphrodite. C and D are shown at the same scale. The scale bar is 250 μm. All animals shown here are progeny of daf-4/+ male and hermaphrodite parents grown at 25° on a single plate. Animals in A and C are either wild type or daf-4/+ heterozygotes.
Curiously, the Cb-daf-4(sa973) strain carrying the Ce-daf-4(+) transgene was not obviously rescued for the Daf-c phenotype at 27° (data not shown). To determine whether this was due to mosaicism, in which not all cells in the animal carried the transgene, we integrated the transgene into the genome (Table 4) (see materials and methods). We found that the integrated lines also did not rescue dauer formation at 27°. Since Ce-daf-4 rescues the Sma phenotype at 27°, this is not explained solely by the inability of CE-DAF-4 to function at high temperatures. We think a more likely explanation is that the lack of rescue reflects a change in the cis-regulatory mechanism of transcriptional regulation and that Ce-daf-4 is not expressed in the cells where Cb-daf-4 functions to control dauer formation. A similar type of change in cis-regulatory mechanisms has been observed for the lin-48 gene, required for positioning of the excretory canal in C. elegans but not C. briggsae (Wang and Chamberlin 2002, 2004).
TABLE 4.
Integrants of saEx[myo-2:gfp, Ce-daf-4(+)]
| Integrant | Linkage |
|---|---|
| syIs802 | X |
| syIs803 | II |
| syIs804 | X |
| syIs807 | IV |
Absence of high temperature-induced dauer formation in C. briggsae:
In C. elegans, in addition to broad temperature sensitivity of the dauer formation process observed over the 15° to 25° temperature range, high temperatures (27° and above) are exceptionally strongly dauer inducing (Ailion and Thomas 2000). Wild-type animals become hypersensitive to dauer pheromone at high temperatures, and also form some dauers in the absence of pheromone. Many mutants that are weakly Daf-c become strongly Daf-c at 27°. In addition, several classes of mutants that are Daf-d at lower temperatures, including daf-3 and cilium structure mutants, become Daf-c at 27°. It is possible that this response in the wild-type C. elegans protects it from temperatures at which they are unable to grow.
To test whether a similar strong induction could be observed in C. briggsae, we looked for dauer formation at high temperatures by the wild type (AF16), two cilium structure mutants [dyf(sa934) and dyf(sy5314)], Cb-daf-3(sy5417), and a weak Daf-c mutant [daf-c(sa974)]. Unlike in C. elegans, the wild type was not observed to form dauers at temperatures above 27°, nor did they form dauers close to the upper temperature limit of growth and reproduction for C. briggsae, 31.5°. In addition, dyf mutants and Cb-daf-3(sy5417) mutants did not form dauers at high temperatures (Figure 3B). This is in contrast to C. elegans where most dyf mutations and daf-3 alleles cause animals to form 100% dauers at 27°. Finally, the weak Daf-c mutant daf-c(sa974) formed relatively few dauers at all temperatures we assayed. This again contrasts with C. elegans, where all known mutants that form some dauers at 25° form 100% dauers at 27°. These results strongly suggest that the specific high-temperature response we observe in C. elegans is not present in C. briggsae.
DISCUSSION
Comparison of the dauer formation pathway in C. elegans and C. briggsae:
Our results demonstrate that the overall pattern of daf gene functions and interactions is conserved between C. elegans and C. briggsae. We isolated mutations in Cb-daf-2, Cb-daf-3, Cb-daf-4, and in genes that affect the formation of sensory cilia. In general, these mutations cause phenotypes very similar to mutations in orthologous C. elegans genes: Cb-daf-3 and cilium structure mutants are Daf-d, whereas Cb-daf-2 and Cb-daf-4 mutants are Daf-c. These similarities extend to their respective pleiotropies. The Cb-daf-2 mutant was long-lived, the Cb-daf-4 mutant was small and exhibited male tail defects, and cilium structure mutants were Dyf. Furthermore, the pattern of interactions among these genes is conserved. In particular, Cb-daf-3(sy5417) suppresses Cb-daf-4(sa973), and Cb-daf-2(sy5445) is epistatic to Cb-daf-3(sy5417). Thus, the pathways regulating dauer formation are largely conserved between C. elegans and C. briggsae.
In contrast to the overall conservation of pathways, C. briggsae and C. elegans differ in their responses to high temperatures. Temperatures above 26° are strongly dauer promoting for C. elegans (Ailion and Thomas 2000). Our examination of C. briggsae wild-type and mutant (both Daf-d and Daf-c) animals at 27° and 31.5° strongly argues against the presence of a similar response in C. briggsae. Related Caenorhabditis species [e.g., C. remanei, C. sp. PS1010, C. brenneri (Sudhaus and Kiontke 2007)] are able to grow at higher temperatures than C. elegans (our data not shown), suggesting that this may be a recently acquired behavioral response in C. elegans. Further molecular analysis of the dauer formation response in C. briggsae and C. elegans potentially can shed light on mechanisms by which neuronally controlled behaviors change during evolution.
Forward genetic analysis in C. briggsae:
Increasingly, C. briggsae is becoming an important model organism for comparative studies. Aspects of C. briggsae biology that are under investigation include positioning of the excretory duct cell (Wang and Chamberlin 2002, 2004), functions of Notch receptors glp-1 and lin-12, (Rudel and Kimble 2001, 2002), male tail development (Baird et al. 2005), vulval development (Delattre and Felix 2001), and molecular evolution of gene families (e.g., Thomas et al. 2005). One particularly fruitful line of investigation examined the mechanism of sex determination. Molecular and phylogenetic analyses indicate that hermaphroditism in C. elegans and C. briggsae evolved independently from male/female ancestors (Cho et al. 2004; Kiontke et al. 2004). Molecular analyses of the sex-determination gene fog-2 and functional analyses of genes including gld-1, fem-2, and fem-3 revealed significant differences in mechanisms by which hermaphrodites are produced in C. elegans and C. briggsae, consistent with convergent, independent evolution of hermaphroditism (Hill et al. 2006; Nayak et al. 2005). Hill et al. (2006) also highlighted an important technical point: although progress can be made with the use of RNAi, because RNAi is often variable and does not fully eliminate gene function, chromosomal null and reduction-of-function mutations are needed to rigorously compare functions of genes in the two species.
Ours is one of the first systematic efforts to generate a large number of mutations affecting a single process in C. briggsae, and our results demonstrate that forward genetic analysis in C. briggsae can be rapid. In general, a mutagenesis and a screen are followed by basic characterizations of isolated alleles. Pleiotropic phenotypes and mapping data provide clues to the molecular identity on the basis of homology to C. elegans, and the identity can be ascertained rapidly by transformation rescue or by direct sequencing of the gene. Once mutations are identified in a few key genes, additional reagents may be generated easily by using preexisting alleles, as demonstrated by our isolation of a Cb-daf-2 allele from a reversion screen of Cb-daf-3. Null alleles, which are necessary for quantitative comparisons of gene functions, can be generated from non-null alleles of the same gene by noncomplementation screens.
A similar conclusion is also true for a nematode more distantly related to C. elegans, Pristionchus pacificus, where a number of mutations from forward genetic analyses have been cloned molecularly (Sommer 2006). C. briggsae is much more closely related to C. elegans and thus shows a greater degree of phenotypic similarity and synteny (Stein et al. 2003). Moreover, the availability of the genome sequence, a genetic map with a large number of visible mutations, and a collection of SNP (single nucleotide polymorphism) markers raise the possibility of easily cloning genes de novo in C. briggsae without relying on homology to C. elegans (Gupta et al. 2007). The closer relationship of C. briggsae to C. elegans means that different types of evolutionary changes will be studied by comparisons of these two species than in comparisons between C. elegans and P. pacificus. Neuronally controlled traits, such as dauer formation, may be more variable between Caenorhabiditis species than morphological traits like vulval development, which is being investigated in C. elegans and P. pacificus (Sommer 2006).
Genetic tools for study of C. briggsae:
Our work on dauer formation generated useful reagents, which will facilitate analysis of gene functions in C. briggsae using mutants. For example, we integrated transgenic arrays containing myo-2∷gfp and Ce-daf-4(+) on chromosomes 2, 4, and X. Since myo-2∷gfp is a bright fluorescent reporter expressed in all stages, these integrants can be used as dominant markers for these chromosomes in mapping experiments and strain constructions. The X-linked myo-2∷gfp integrant syIs802 has also proved to be useful because syIs802/+ heterozygotes produce high frequency of XO male progeny, probably by interfering with meiotic pairing (Hill et al. 2006). Finally, molecularly characterized mutations with easily scored phenotypes contribute to increasing the density of the C. briggsae genetic map.
Acknowledgments
We thank R. Johnsen and D. Baillie for C. briggsae mutants and mapping data. We thank S. Gharib and B. P. Gupta for technical assistance and useful reagents. We thank E. Hallem, X. Wang, and J. Srinivasan for comments on the manuscript. T.I. was supported by fellowship DRG-1646 from the Damon Runyon Cancer Research Foundation. This work was supported by HHMI, where P.W.S. is an investigator. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources (NCRR).
References
- Ailion, M., and J. H. Thomas, 2000. Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics 156: 1047–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi, A., W. H. Yeh, D. Tait, E. M. Hedgecock and D. L. Riddle, 2000. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 14: 1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Baird, S. E., C. R. Davidson and J. C. Bohrer, 2005. The genetics of ray pattern variation in Caenorhabditis briggsae. BMC Evol. Biol. 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnby, D. A., E. M. Link, J. J. Vowels, H. Tian, P. L. Colacurcio et al., 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics 155: 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher, R. A., M. Fujita, F. C. Schroeder and J. Clardy, 2007. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat. Chem. Biol. 3: 420–422. [DOI] [PubMed] [Google Scholar]
- Cassada, R. C., and R. L. Russell, 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46: 326–342. [DOI] [PubMed] [Google Scholar]
- Cho, S., S. W. Jin, A. Cohen and R. E. Ellis, 2004. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 14: 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., M. A. Felix, A. Barriere and D. Charlesworth, 2006. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173: 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Graca, L. S., K. K. Zimmerman, M. C. Mitchell, M. Kozhan-Gorodetska, K. Sekiewicz et al., 2004. DAF-5 is a Ski oncoprotein homolog that functions in a neuronal TGF beta pathway to regulate C. elegans dauer development. Development 131: 435–446. [DOI] [PubMed] [Google Scholar]
- Delattre, M., and M. A. Felix, 2001. Polymorphism and evolution of vulval precursor cell lineages within two nematode genera, Caenorhabditis and Oscheius. Curr. Biol. 11: 631–643. [DOI] [PubMed] [Google Scholar]
- Estevez, M., L. Attisano, J. L. Wrana, P. S. Albert, J. Massague et al., 1993. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365: 644–649. [DOI] [PubMed] [Google Scholar]
- Fodor, A., D. L. Riddle, F. K. Nelson and J. W. Golden, 1983. Comparison of a new wild type Caenorhabditis briggsae with laboratory strains of C. briggsae and C. elegans. Nematologica 29: 203–217. [Google Scholar]
- Gil, E. B., E. Malone Link, L. X. Liu, C. D. Johnson and J. A. Lees, 1999. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc. Natl. Acad. Sci. USA 96: 2925–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, J. W., and D. L. Riddle, 1982. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218: 578–580. [DOI] [PubMed] [Google Scholar]
- Golden, J. W., and D. L. Riddle, 1984. a A pheromone-induced developmental switch in Caenorhabditis elegans: Temperature-sensitive mutants reveal a wild-type temperature-dependent process. Proc. Natl. Acad. Sci. USA 81: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, J. W., and D. L. Riddle, 1984. b The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev. Biol. 102: 368–378. [DOI] [PubMed] [Google Scholar]
- Gupta, B. P., R. Johnsen and N. Chen, 2007. Genomics and biology of the nematode Caenorhabditis briggsae (May 3, 2007), in WormBook, edited by The C. elegans Research Community (http://www.wormbook.org). [DOI] [PMC free article] [PubMed]
- Hill, R. C., C. E. de Carvalho, J. Salogiannis, B. Schlager, D. Pilgrim et al., 2006. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev. Cell 10: 531–538. [DOI] [PubMed] [Google Scholar]
- Hillier, L. W., R. D. Miller, S. E. Baird, A. Chinwalla, L. A. Fulton et al., 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., 1995. Caenorhabditis elegans. Trends Genet. Genetic Nomenclature Guide, 24–25. [PubMed]
- Horvitz, H. R., S. Brenner, J. Hodgkin and R. K. Herman, 1979. A uniform genetic nomenclature for the nematode Caenorhabditis elegans. Mol. Gen. Genet. 175: 129–133. [DOI] [PubMed] [Google Scholar]
- Jeong, P.-Y., M. Jung, Y.-H. Yim, H. Kim, M. Park et al., 2005. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 433: 541–545. [DOI] [PubMed] [Google Scholar]
- Kennedy, B. P., E. J. Aamodt, F. L. Allen, M. A. Chung, M. F. Heschl et al., 1993. The gut esterase gene (ges-1) from the nematodes Caenorhabditis elegans and Caenorhabditis briggsae. J. Mol. Biol. 229: 890–908. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., J. Chang, E. Gensch, A. Rudner and R. Tabtiang, 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Kimura, K. D., H. A. Tissenbaum, Y. Liu and G. Ruvkun, 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946. [DOI] [PubMed] [Google Scholar]
- Kiontke, K., N. P. Gavin, Y. Raynes, C. Roehrig, F. Piano et al., 2004. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101: 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac, M., and P. W. Sternberg, 2003. cis-Regulatory control of three cell fate-specific genes in vulval organogenesis of Caenorhabditis elegans and C. briggsae. Dev. Biol. 257: 85–103. [DOI] [PubMed] [Google Scholar]
- Lin, K., J. B. Dorman, A. Rodan and C. Kenyon, 1997. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322. [DOI] [PubMed] [Google Scholar]
- Malone, E. A., and J. H. Thomas, 1994. A screen for nonconditional dauer-constitutive mutations in Caenorhabditis elegans. Genetics 136: 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova, V. T., C. Z. Borland, L. Manjarrez, M. J. Stern and H. Sun, 1999. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc. Natl. Acad. Sci. USA 96: 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J. Z., H. A. Tissenbaum and G. Ruvkun, 1996. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382: 536–539. [DOI] [PubMed] [Google Scholar]
- Nayak, S., J. Goree and T. Schedl, 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg, S., and G. Ruvkun, 1998. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell 2: 887–893. [DOI] [PubMed] [Google Scholar]
- Ogg, S., S. Paradis, S. Gottlieb, G. I. Patterson, L. Lee et al., 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999. [DOI] [PubMed] [Google Scholar]
- Paradis, S., M. Ailion, A. Toker, J. H. Thomas and G. Ruvkun, 1999. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 13: 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G. I., A. Koweek, A. Wong, Y. Liu and G. Ruvkun, 1997. The DAF-3 Smad protein antagonizes TGF-beta-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes Dev. 11: 2679–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, L. A., E. M. Hedgecock, J. N. Thomson and J. G. Culotti, 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117: 456–487. [DOI] [PubMed] [Google Scholar]
- Prasad, B. C., B. Ye, R. Zackhary, K. Schrader, G. Seydoux et al., 1998. unc-3, a gene required for axonal guidance in Caenorhabditis elegans, encodes a member of the O/E family of transcription factors. Development 125: 1561–1568. [DOI] [PubMed] [Google Scholar]
- Riddle, D. L., 1977. A genetic pathway for dauer larva formation in Caenorhabditis elegans. Stadler Genet. Symp. 9: 101–120. [Google Scholar]
- Riddle, D. L., M. M. Swanson and P. S. Albert, 1981. Interacting genes in nematode dauer larva formation. Nature 290: 668–671. [DOI] [PubMed] [Google Scholar]
- Rouault, J. P., P. E. Kuwabara, O. M. Sinilnikova, L. Duret, D. Thierry-Mieg et al., 1999. Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr. Biol. 9: 329–332. [DOI] [PubMed] [Google Scholar]
- Rudel, D., and J. Kimble, 2001. Conservation of glp-1 regulation and function in nematodes. Genetics 157: 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel, D., and J. Kimble, 2002. Evolution of discrete Notch-like receptors from a distant gene duplication in Caenorhabditis. Evol. Dev. 4: 319–333. [DOI] [PubMed] [Google Scholar]
- Savage, C., P. Das, A. L. Finelli, S. R. Townsend, C. Y. Sun et al., 1996. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc. Natl. Acad. Sci. USA 93: 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, R. J., 2006. Pristionchus pacificus (August 14, 2006), WormBook, edited by The C. elegans Research Community (http://www.wormbook.org). [DOI] [PMC free article] [PubMed]
- Starich, T. A., R. K. Herman, C. K. Kari, W. H. Yeh, W. S. Schackwitz et al., 1995. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139: 171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L. D., Z. Bao, D. Blasiar, T. Blumenthal, M. R. Brent et al., 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhaus, W., and K. Kiontke, 2007. Comparison of the cryptic nematode species Caenorhabditis brenneri sp. n. and C. remanei (Nematoda: Rhabditidae) with the stem species pattern of the Caenorhabditis Elegans group. Zootaxa 1456: 45–62. [Google Scholar]
- Sulston, J. E., and H. R. Horvitz, 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Swoboda, P., H. T. Adler and J. H. Thomas, 2000. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell 5: 411–421. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., 1993. Chemosensory regulation of development in C. elegans. Bioessays 15: 791–797. [DOI] [PubMed] [Google Scholar]
- Thomas, J. H., D. A. Birnby and J. J. Vowels, 1993. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134: 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J. H., J. L. Kelley, H. M. Robertson, K. Ly and W. J. Swanson, 2005. Adaptive evolution in the SRZ chemoreceptor families of Caenorhabditis elegans and Caenorhabditis briggsae. Proc. Natl. Acad. Sci. USA 102: 4476–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels, J. J., and J. H. Thomas, 1992. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130: 105–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and H. M. Chamberlin, 2002. Multiple regulatory changes contribute to the evolution of the Caenorhabditis lin-48 ovo gene. Genes Dev. 16: 2345–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and H. M. Chamberlin, 2004. Evolutionary innovation of the excretory system in Caenorhabditis elegans. Nat. Genet. 36: 231–232. [DOI] [PubMed] [Google Scholar]
- Way, J. C., and M. Chalfie, 1989. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 3: 1823–1833. [DOI] [PubMed] [Google Scholar]