Abstract
Mitochondrial tRNA import is widespread in eukaryotes. Yet, the mechanism that determines its specificity is unknown. Previous in vivo experiments using the tRNAsMet, tRNAIle and tRNALys have suggested that the T-stem nucleotide pair 51:63 is the main localization determinant of tRNAs in Trypanosoma brucei. In the cytosol-specific initiator tRNAMet, this nucleotide pair is identical to the main antideterminant that prevents interaction with cytosolic elongation factor (eEF1a). Here we show that ablation of cytosolic eEF1a, but not of initiation factor 2, inhibits mitochondrial import of newly synthesized tRNAs well before translation or growth is affected. tRNASec is the only other cytosol-specific tRNA in T. brucei. It has its own elongation factor and does not bind eEF1a. However, a mutant of the tRNASec expected to bind to eEF1a is imported into mitochondria. This import requires eEF1a and aminoacylation of the tRNA. Thus, for a tRNA to be imported into the mitochondrion of T. brucei, it needs to bind eEF1a, and it is this interaction that mediates the import specificity.
Keywords: elongation factor1a, mitochondrial biogenesis, selenocysteine tRNA, tRNA, trypanosomes
Introduction
Most protozoa, many fungi, plants and a few animals lack a variable number of mitochondrial tRNA genes. It has been shown in these organisms that the missing genes are compensated for by import of a small fraction of the corresponding cytosolic tRNAs (Schneider and Marechal-Drouard, 2000; Bhattacharyya and Adhya, 2004). The phylogenetic distribution of mitochondrial tRNA import is disperse. Thus, for some species where tRNA import has been predicted, closely related organisms can be found that do not import tRNAs (Schneider and Marechal-Drouard, 2000). Since the loss of mitochondrial tRNA genes is likely to be irreversible, this suggests that the process has a polyphyletic origin. This conclusion is supported by studies of mitochondrial tRNA import in yeast (Tarassov et al, 1995), Leishmania (Goswami et al, 2006) and plants (Salinas et al, 2006), which provided evidence for three distinct tRNA import machineries. The capability to import tRNAs in these three groups of organisms is therefore due to convergent evolution.
Consistent with this view is the fact that the number of imported tRNAs is species-specific. Mitochondria of Saccharomyces cerevisiae import two tRNAs only (Tarassov and Martin, 1996; Rinehart et al, 2005). Plants import a variable number of mitochondrial tRNAs, but have retained at least a few mitochondrial tRNA genes (Dietrich et al, 1996b). The most extreme cases are two groups of unrelated parasitic protozoa, the trypanosomatids (which include Trypanosoma brucei and Leishmania spp.) (Simpson et al, 1989; Hancock and Hajduk, 1990; Schneider et al, 1994) and the apicomplexans (Crausaz-Esseiva et al, 2004b), both of which completely lack mitochondrial tRNA genes and therefore must import the whole set of tRNAs. However, in both parasites we still find tRNAs that are cytosol-specific (Crausaz-Esseiva et al, 2004a, 2004b; Geslain et al, 2006). Interestingly, in all organisms that have been analyzed, an imported nucleus-encoded mitochondrial tRNA only represents a small fraction of a normal cytosolic tRNA (Schneider and Marechal-Drouard, 2000; Tan et al, 2002b). Strikingly, the imported fraction is specific for a given tRNA species and varies between 1 and 8%.
Thus, two prominent questions regarding mitochondrial targeting of tRNAs are (i) what determines the import specificity and (ii) what regulates the extent of tRNA import? Regarding the latter, it has been suggested that for some leishmanial tRNAs the extent of import is regulated by cytosol-specific thio-modifications in the anticodon (Kaneko et al, 2003). Regarding the former, there are a number of studies in different organisms showing that the import specificity is controlled by localization determinants on mature tRNAs (Rusconi and Cech, 1996; Entelis et al, 1998; Crausaz-Esseiva et al, 2004a). However, as expected due to the polyphyletic origin of tRNA import, they are not identical in the different species. In the imported tRNALys isoacceptor of yeast, the localization signals are confined to the acceptor stem and the anticodon loop, and are required for binding to the precursor of mitochondrial lysyl-tRNA synthetase (Entelis et al, 1998). This protein forms a complex with the imported tRNALys, which then is transported across the mitochondrial membranes by using the protein import pores (Tarassov et al, 1995). It is not known how the other imported yeast tRNA, the tRNAGln, is addressed to mitochondria, and by which mechanism it is imported (Rinehart et al, 2005).
The only other species where the in vivo tRNA import determinants have been analyzed in detail are Tetrahymena and T. brucei. For tRNAGln isoacceptors of Tetrahymena it is the anticodon (Rusconi and Cech, 1996), and for the tRNAMet isoacceptors of T. brucei a single T-stem nucleotide pair that are both necessary and sufficient to determine the localization of these tRNAs (Crausaz-Esseiva et al, 2004a). Only fragmentary results are available for what determines the in vivo import specificity in plants; a point mutation in the acceptor stem of tRNAAla of potato was shown to abolish import in vivo (Dietrich et al, 1996a), and more recently the D-loop and the anticodon region were implicated in import of plant tRNAVal (Delage et al, 2003).
However, it is not known in any system which factors decode the localization signals. Here we present evidence that in T. brucei, binding to translation elongation factor 1a (eEF1a) is a prerequiste for import, suggesting that it is this interaction that determines the specificity of tRNA import in vivo.
Results
Correlation between import and binding to EF1a
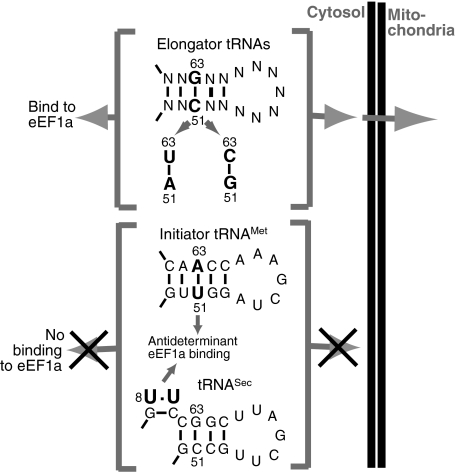
In T. brucei the initiator tRNAMet (Crausaz-Esseiva et al, 2004a) and the tRNASec are cytosol-specific (Geslain et al, 2006). All other tRNAs function in both the cytosol and the mitochondrion (Figure 1). Thus, by expressing chimeras between the closely related cytosolic initiator and the imported elongator tRNAsMet, we showed that the single unmodified T-stem nucleotide pair at position 51:63 is both necessary and sufficient for the correct localization of the tRNAsMet (Crausaz-Esseiva et al, 2004a). The adjacent nucleotide pair 52:62 influences the efficiency of import, but when transplanted onto other tRNAs, was not able to change their localization. Furthermore, we showed that both the cytosolic as well as the mitochondrial localization determinants can act in the context of the tRNAIle and the tRNALys (Crausaz-Esseiva et al, 2004a), suggesting that the same determinants can function in the context of any trypanosomal tRNA. Thus, if we find the T-stem nucleotide pair U51:A63, the tRNA remains in the cytosol, whereas if any other standard base pair, such as C:G, A:U or G:C, is present at this position, the tRNA is in part imported into mitochondria (Crausaz-Esseiva et al, 2004a) (Figure 1). (However, the tRNASec is an exception, despite carrying C51:G63 it is cytosol-specific.) Interestingly, the nucleotide pair U51:A63 is conserved in all eukaryotic initiator tRNAsMet and generally absent from elongator tRNAs. It not only acts as a cytosolic localization determinant in T. brucei, but the corresponding nucleotide pair in vertebrate initiator tRNAMet is one of two antideterminants that prevent binding of cytosolic eEF1a (Drabkin et al, 1998). The trypanosomal eEF1a is 78% identical to its human counterpart (Kaur and Ruben, 1994), which makes it very likely that the U51:A63 nucleotide pair also acts as antideterminant for the T. brucei protein. Furthermore, it has been shown that one tRNA domain recognized by eEF1a is the T-arm (Dreher et al, 1999). Thus, we observe a perfect correlation between mitochondrial import of a given trypanosomal tRNA and its predicted binding to eEF1a. This is not only true for wild-type tRNAs but also for the numerous variants whose localization we have tested in vivo (Crausaz-Esseiva et al, 2004a). In agreement with this correlation we see a congruence of the localization determinant with a nucleotide pair involved in binding or preventing of binding to eEF1a. Based on these observations we suggest the hypothesis that in T. brucei interaction with eEF1a is a prerequisite for a tRNA to be imported into mitochondria, and that it is this binding that determines the specificity of the process.
Figure 1.
Specificity of mitochondrial tRNA import in T. brucei. Top part: All elongator tRNAs specifying the 20 standard amino acids are in part imported into mitochondria. The signal that determines their localization is the T-stem nucleotide pair C51:63G, A51:U63 or G51:C63. Lower part: The initiator tRNAMet and the tRNASec are cytosol-specific. The cytosolic localization signal of initiator tRNAMet, the nucleotide pair U51:A63 (Crausaz-Esseiva et al, 2004a), is at the same time the major antideterminant for eEF1a binding (Drabkin et al, 1998). The T-stem loop region of the cytosolic tRNASec includes a putative C51:G63 import signal. However, the non-standard U:U nucleotide pair (number 8 in the acceptor stem) is a putative antideterminant for eEF1a binding (Rudinger et al, 1996). Thus, all tRNAs that interact with eEF1a are imported, whereas the ones that do not are cytosol-specific.
How does the cytosolic localization of the tRNASec, which lacks the U51:A63 cytosolic localization determinant of the initiator tRNAMet, fit into this picture (Figure 1)? It is known that tRNAsSec do not bind to eEF1a (or the bacterial homologue EF-Tu). In eukaryotes this is most likely due to the non-conventional U:U nucleotide pair at position 9 of the acceptor stem, which acts as an antideterminant for eEF1a binding (Rudinger et al, 1996). tRNAsSec, instead of eEF1a, interact with their own specialized elongation factor, termed EFSec (Diamond, 2004), an orthologue of which has also been identified in T. brucei (Cassago et al, 2006; Lobanov et al, 2006). Taking all this into account, the cytosolic localization of the tRNASec, rather than contradicting our hypothesis, actually supports it.
Inducible tRNA expression
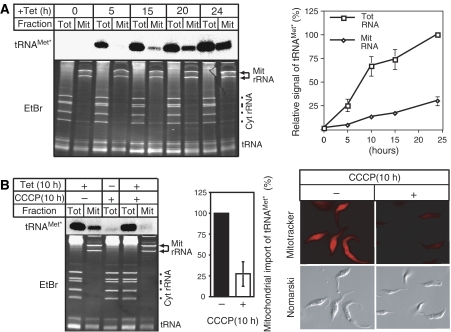
In order to test the hypothesis that eEF1a is involved in tRNA import, we constructed RNAi cell lines allowing inducible ablation of either eEF1a or as a control of cytosolic translation initiation factor 2 (eIF2). Ablation of both of these proteins, as expected due to their essential function in translation, leads to a growth arrest but did not change the steady-state levels of mitochondrial tRNAs (data not shown). This could however be due to the fact that even in the absence of import the tRNA population that was imported before the induction of RNAi may persist for a long time. It might therefore not be possible to detect an import phenotype by simply analyzing the steady-state population of tRNAs. The very same problem was encountered in the analysis of mitochondrial protein import in yeast, where inducible ablation of a key import factor did not result in an obvious depletion of mitochondrial-localized proteins at steady state (Baker et al, 1990). However, the import phenotype was clearly seen in a pulse–chase experiment, which allows to selectively monitor newly synthesized proteins. Thus, in order to follow the fate of a newly synthesized tRNA in T. brucei, we produced a cell line that allows inducible expression of a nucleus-encoded and imported tRNA. Practically this was achieved by transfection of T. brucei 29-13, which expresses the tetracycline repressor, with a construct containing the tetracycline operator 5′ of a tagged tRNA gene. Figure 2A shows that in these cells addition of tetracycline induces expression of the tagged tRNA in a time-dependent manner. The transgenic tRNA is correctly processed as well as aminoacylated (not shown), and by all means behaves like a fully functional tRNA. Analysis of digitonin-extracted mitochondrial fractions furthermore showed that, as expected, the tagged tRNA was imported into mitochondria. In vitro experiments from different laboratories suggested that tRNA import requires an electrochemical gradient across the mitochondrial inner membrane (Mukherjee et al, 1999; Yermovsky-Kammerer and Hajduk, 1999). The left and the middle panels of Figure 2B show that treatment of a culture of T. brucei with carbonyl cyanide m-chlorophenylhydrazone (CCCP)—an uncoupler that dissipates the electrochemical gradient—inhibits import of the newly synthesized tRNA by 75%. This inhibition is not detected by looking at the steady-state mitochondrial tRNA pool (Figure 2B, left panel), since the major fraction of each tRNA was imported before the CCCP treatment. Staining of cells with Mitotracker (Figure 2B, right panel), a dye that detects the electrochemical gradient, confirms that incubation with CCCP depolarizes the mitochondrial inner membrane and shows that the cells remain alive and morphologically unchanged during the treatment.
Figure 2.
Tetracycline-inducible expression of a tagged tRNA. (A) Time course of induction. Appearance of the tagged tRNAMet (tRNAMet*) in the cytosol (Tot) and in digitonin-extracted mitochondria (Mit) was monitored by Northern analysis (left side, upper panel). The lower panel shows the corresponding ethidium bromide-stained gel (EtBr). Positions of the mitochondrial rRNAs (Mit rRNA) and the cytosolic rRNAs (Cyt rRNA), as well as the tRNA region are indicated. Graph: Quantitative analysis of four independent experiments of the type shown on the left. The signal corresponding to the tagged tRNAMet at 24 h of induction in the total RNA fraction was set to 100%. Standard errors are indicated. (B) Mitochondrial import of newly synthesized tagged tRNAMet requires the membrane potential. Left panel: Expression of the tagged tRNA was induced for 10 h in absence (−) and presence (+) of 20 mmol of the uncoupler CCCP, and analyzed by Northern blot. Middle panel: Quantitative analysis of four independent experiments of the type shown on the left. The signal in untreated cells that corresponds to the mitochondrially localized tagged tRNAMet after 10 h of induction was set to 100%. Standard errors are indicated. Right panel: Mitotracker-staining of untreated (−) and CCCP-treated cells (+). The y-axis images of the ethidium bromide-stained gels have been electronically compressed by a factor of approximately 2.
Thus, these results demonstrate that in vivo import of trypanosomal tRNAs requires an electrochemical gradient across the inner mitochondrial membrane, and provide a proof of principle that inducible tRNA expression can be used to study aspects of mitochondrial tRNA import that previously were not accessible to direct in vivo analysis.
Inducible tRNA expression combined with RNAi
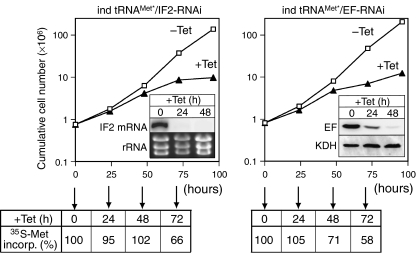
In a next step we produced two RNAi cell lines, which upon addition of tetracycline, induce the expression of the tagged tRNA gene, and at the same time downregulate the expression of eIF2 or eEF1a, respectively. Both cell lines showed a slow growth phenotype approximately 48 h after induction of RNAi (Figure 3). Furthermore, in both cases, concomitant with the growth arrest a reduction of cytosolic protein synthesis as measured by 35S-methionine incorporation was seen (Figure 3).
Figure 3.
Inducible tRNA expression combined with RNAi. Growth curve of a representative clonal T. brucei RNAi cell line allowing simultaneously inducible expression of the tagged tRNAMet and ablation of IF2 (ind tRNAMet*/IF2-RNAi) and eEF1a (ind tRNAMet*/EF-RNAi), respectively. Open squares and filled triangles represent growth in the absence or presence of tetracycline, respectively. The inset in the left graph shows a Northern blot for the eIF2 mRNA (IF2). The rRNAs in the lower panel serve as loading controls. The inset in the right panel shows an immunoblot probed for eEF1a (EF), and as control for α-ketoglutarate dehydrogenase (KDH), which is not affected by the RNAi. Quantitation of the signals illustrates that the RNAi causes efficient ablation of eEF1a relative to KDH, reaching 31 and 5% after 24 and 48 h, respectively. The efficiency of cytosolic translation during induction of RNAi expressed by the percentage of 35S-labeled methionine incorporation into total cellular protein is indicated at the bottom of each graph. 35S-labeled methionine incorporation in uninduced cells was set to 100%.
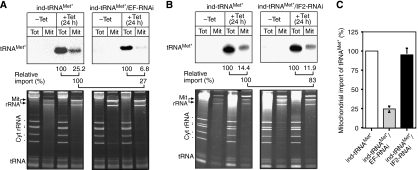
However, fractionation of the eEF1a-ablated cell line showed that reproducibly approximately fourfold less of the newly synthesized tRNA was found in the mitochondrial fraction than in uninduced cells (Figure 4A). In contrast, no significant effect on import of the newly synthesized tRNA was detected in cells ablated for eIF2 (Figure 4B). Most importantly, the tRNA import phenotype in the eEF1a cell line is already detected 24 h after induction of RNAi, well before growth or translation is affected (Figure 3). Consistent with this observation, the induced cells are fully motile and the electrochemical gradient of their mitochondria, as evidenced by Mitotracker staining, is identical to the one observed in uninduced cells (data not shown). As a further control, that it is indeed the lack of eEF1a that causes the import phenotype, we tested the import of the newly synthesized tRNA in a cell line ablated for the seryl-tRNA synthetase, an essential protein that as eIF2 and eEF1a is required for translation. It was previously shown that ablation of this enzyme causes deacylation of tRNAsSer and leads to a growth arrest (Supplementary Figure 1A) whose kinetics is identical to the one seen in the eIF2 and eEF1a RNAi cell lines (Geslain et al, 2006) (Figure 3). According to our model we expect that ablation of the seryl-tRNA synthetase, identical to the knockdown of eIF2 but in contrast to the ablation of eEF1a, will not affect the import of the newly synthesized tRNA, which is exactly what is seen (Supplementary Figure 1B).
Figure 4.
Effect of RNAi on import of newly synthesized tRNA. (A) Presence of the tagged tRNAMet (tRNAMet*) in the cytosol (Tot) and in digitonin-extracted mitochondria (Mit) was monitored by Northern analysis (top panels) in cell lines allowing either inducible expression of the tagged tRNAMet only (ind-tRNAMet*, left panel), or inducible expression of the tagged tRNAMet in combination with ablation of eEF1a (ind tRNAMet*/EF-RNAi, right panel). Relative import efficiency of the newly synthesized tRNAMet are indicated at the bottom. The signal in the total RNA fractions (first line) or the mitochondrial fractions in the control cells (second line) was set to 100%. The lower panels show the corresponding ethidium bromide-stained gels. Positions of the mitochondrial rRNAs (Mit rRNA) and the cytosolic rRNAs (Cyt rRNA), as well as the tRNA region are indicated. (B) Same as panel A, but analysis on the right panel was performed with the cell lines allowing inducible expression of the tagged tRNAMet, in combination with ablation of eIF2 (ind tRNAMet*/IF2-RNAi, right panel). (C) Graph of three independent replicate experiments showing the relative import efficiencies of the newly synthesized tRNA in cells that do not undergo RNAi and in eEF1a and eIF2 ablated cell lines, respectively. Bars=s.e.
These results strongly suggest that inhibition of import of newly synthesized tRNAs is a direct consequence of the lack of eEF1a. However, it cannot formally be excluded that the lack of a labile factor, required for tRNA import, that is rapidly degraded under limiting eEF1a concentrations is responsible for inhibition of import.
To be imported into mitochondria tRNAs must cross both the nuclear and the mitochondrial membranes. A potential caveat of the in vivo import system is to distinguish nuclear retention from inhibition of mitochondrial import. There are two ways to export tRNAs from the nucleus: the exportin-t and the exportin-5 pathway (Bohnsack et al, 2002; Calado et al, 2002). There is no reason to believe that ablation of eEF1a will affect the exportin-t pathway. At first sight this looks different for the exportin-5 pathway, since it transports both tRNAs and eEF1a. However, nuclear export of eEF1a requires the presence of tRNAs that bind to both eEF1a and exportin-5. Thus, while export of eEF1a depends on tRNAs, the converse is not true and tRNAs are still exported even in the absence of eEF1a (Bohnsack et al, 2002; Calado et al, 2002). Finally, we have addressed this question experimentally for the tRNASec variants that are discussed in the next section.
In summary, inhibition of tRNA import by ablation of EF1a shows that in T. brucei eEF1a has a dual function; besides its role in cytosolic translation, it is required for in vivo import of tRNAs into mitochondria and determines the specificity of the process.
The tRNASec
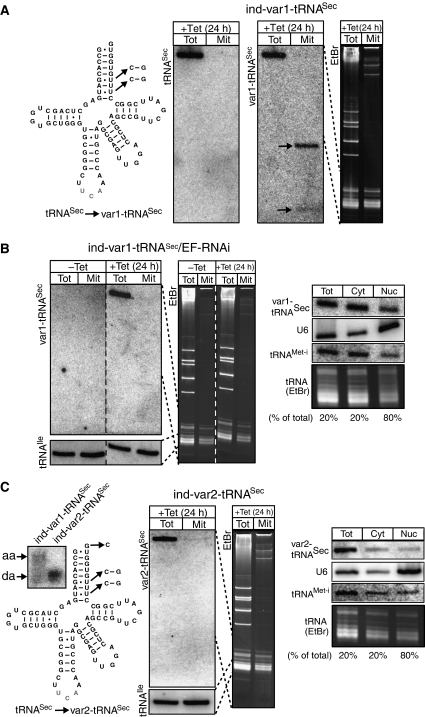
Eukaryotic and bacterial tRNAsSec do not interact with eEF1a or EF-Tu, respectively (Diamond, 2004). Instead they have their own elongation factors. The cytosolic localization of the trypanosomal tRNASec therefore supports the hypothesis that binding to eEF1a might be a prerequisite for tRNA import. For Escherichia coli tRNASec, the antideterminants for EF-Tu binding have been mapped to the eighth, ninth and tenth base pairs of the acceptor branch (Rudinger et al, 1996). Interestingly, the eighth acceptor stem base pair of eukaryotic tRNASec is invariantly a non-Watson Crick U:U (Figure 5A). It has been suggested, in analogy to the situation in bacteria, that this base pair may act as an antideterminant for eEF1a binding in eukaryotes (Rudinger et al, 1996). Thus, we would expect that a variant of the trypanosomal tRNASec, where the U:U eEF1a antideterminant had been replaced by a standard C:G base pair should bind eEF1a (Figure 5A). Our hypothesis predicts that as a consequence this variant tRNASec should be imported into mitochondria. In transgenic T. brucei cells that express the variant tRNASec, this is indeed observed and sequences derived from the variant tRNASec, contrary to the wild-type tRNASec, are recovered in both the cytosol and the mitochondrial fraction (Figure 5A). However, instead of the intact molecule we reproducibly detect two distinct smaller fragments. Thus, for unknown reasons the tRNASec variant appears to be degraded when present in mitochondria.
Figure 5.
Mitochondrial import of a tRNASec variants. (A) Predicted secondary structure of the trypanosomal tRNASec. The nucleotide changes that were introduced to obtain the variant tRNASec (var1-tRNASec) that lacks the predicted eEF1a antideterminant are indicated. RNA from the cytosol (Tot) and from digitonin-extracted mitochondria (Mit) of a cell line allowing tetracycline-inducible expression of the variant tRNASec (ind-var1-tRNASec) was analyzed by specific oligonucleotide hybridization for the presence of the wild-type tRNASec (tRNASec) (left panel) and the variant tRNASec (var1-tRNASec) (middle panel). Arrows highlight the two fragments of the variant tRNASec that are reproducibly detected in the mitochondrial fraction. Right panel: Ethidium bromide staining (EtBr) of the corresponding gel. Broken lines indicate which region of the stained gel is represented in the blot. (B) Effect of eEF1a-RNAi on import of newly synthesized var1-tRNASec. Left panel: Northern analysis for var1-tRNASec of cytosolic (Tot) and digitonin-extracted mitochondrial (Mit) RNA fractions of an uninduced (−Tet) and induced (+Tet) cell line that allows tetracycline-regulated expression of the var1-tRNASec in combination with ablation of eEF1a (var1-tRNASec/EF-RNAi). The growth phenotype of this cell line was essentially identical to the one shown for the eEF1a-ablated cell line shown in Figure 3 (data not shown). Bottom panels show a reprobing of the same blot for the endogenous imported tRNAIle. Middle panel: EtBr staining of the corresponding gel. Broken lines indicate which region of the stained gel correspond to which blots. Right panel: Total (Tot), cytosolic (Cyt) and nuclear (Nuc) RNA fractions were analyzed for the presence of var1-tRNASec, the primarily nuclearly localized U6 RNA (U6), the cytosolic initiator tRNAMet (tRNAMet−i) and for tRNAs in general (tRNAs, EtBr). The percentage of the total samples that were analyzed in the different lanes is indicated at the bottom. (C) Predicted secondary structure of the tRNASec. The discriminator nucleotide change that prevents charging by seryl-tRNA synthetase and the nucleotide changes that inactivate the predicted eEF1a antideterminant are indicated. All these changes lead to a variant tRNASec that is termed var2-tRNASec. Left panel: Total RNA from cell lines allowing tetracycline-inducible expression of the var1-tRNASec (ind-var1-tRNASec) and var2-tRNASec (ind-var2-tRNASec), respectively, was analyzed on a long acidic gel. Aminoacylated (aa) and deacylated (da) var1-tRNASec (left lane) and var2-tRNASec (right lane) were detected by specific oligonucleotide hybridization. Middle two panels: RNA from the cytosol (Tot) and from digitonin-extracted mitochondria (Mit) of the var2-tRNASec expressing cell line was analyzed for the presence var2-tRNASec. The corresponding EtBr-stained gel is also shown. Broken lines indicate which region of the stained gel corresponds to which blot. Right panel: Distribution of var2-tRNASec in total, cytosolic and nuclear RNA fractions (as in (B)).
In a next experiment we prepared a cell line allowing inducible expression of the tRNASec variant with simultaneous knockdown of eEF1a (Figure 5B). Induction of RNAi led to a similar growth phenotype than is observed in the previously described eEF1a RNAi cell line (Figure 3) (data not shown). As expected according to our model, ablation of eEF1a for 24 h abolished mitochondrial import of the variant tRNASec (Figure 5B, left panel).
In order to show that the variant tRNASec accumulates in the cytosol and not in the nucleus, we performed cell fractionations using the detergent digitonin. A quantification of the lanes in the right panel of Figure 5B shows—after normalization to equal cell equivalents—that 50% of the primarily nucleus-localized U6 RNA is recovered in the pellet. However, 88% each of the cytosolic initiator tRNAMet and the variant tRNASec are recovered in the supernatant, confirming that ablation of eEF1a does not interfere with nuclear tRNA export (Figure 5B, right panel).
Thus, these experiments directly link import of the variant tRNASec to the presence of eEF1a.
Formation of the ternary complex between eEF1a, GTP and tRNA requires the tRNA to be aminoacylated (Ribeiro et al, 1995). We have recently shown that the discriminator nucleotide G73 on tRNASer and the tRNASec is the major identity element recognized by the trypanosomal seryl-tRNA synthetase (Geslain et al, 2006). Thus, changing the G73 on the tRNASec to a C is expected to abolish aminoacylation. The Northern blot in the left panel of Figure 5C shows that the same is true for the tRNASec variant that is imported into mitochondria. Interestingly, cell fractionation reveals that this aminoacylation-deficient tRNASec variant cannot anymore be imported into mitochondria (Figure 5C, middle two panels) even though it lacks—just as the imported variant in Figure 5A—the antideterminant for eEF1a binding. It is important to emphasize that this experiment is not based on RNAi. Translation is therefore fully active. Quantification of a cell fractionation experiment (Figure 5C, right panel) shows that the absence of mitochondrial import of the variant tRNASec cannot be explained by nuclear retention of the aminoacylation-deficient tRNASec. In this experiment 50% of the U6 RNA is found in the nuclear fraction, whereas 81% of the cytosolic initiator tRNAMet and 88% of the variant tRNASec are recovered in the cytosol.
Thus, the most parsimonious explanation for these results is that in absence of aminoacylation the tRNA cannot bind to eEF1a and therefore is not imported into mitochondria.
In summary, eEF1a-dependent mitochondrial import of the tRNASec variant that is predicted to interact with eEF1a, strongly supports the notion that this protein is required for mitochondrial tRNA import in T. brucei.
In vitro import shows no specificity
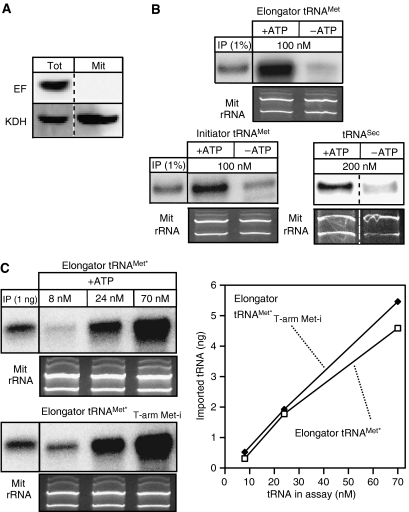
A number of groups have shown that efficient import of tRNAs into isolated mitochondria of trypanosomatids occurs in the absence of added cytosol and thus of eEF1a (Mukherjee et al, 1999; Yermovsky-Kammerer and Hajduk, 1999; Rubio et al, 2000; Crausaz-Esseiva et al, 2004a). This raises the question of how these results can be reconciled with the eEF1a-dependent targeting mechanism proposed in our model? Should the specificity of tRNA import indeed be mediated by eEF1a, we would predict that in vitro all tRNAs, irrespective of whether in vivo they are cytosol-specific or not, should be imported. In order to test this prediction, we performed in vitro import experiments using transcripts corresponding to the in part imported elongator tRNAMet as well as to the cytosol-specific initiators tRNAMet and the tRNASec. Figure 6 shows that all these cytosol-specific tRNAs are imported into isolated mitochondria that are devoid of eEF1a. The import efficiency of the initiator tRNAMet is comparable to that of the elongator tRNAMet, which in vivo is partly localized to the mitochondria (Crausaz-Esseiva et al, 2004a). Figure 6C indicates that the import efficiency does not depend on the substrate concentration. Both elongator tRNAMet and a variant thereof, carrying the T-stem of the in vivo cytosolically localized initiator tRNAMet (Crausaz-Esseiva et al, 2004a), were imported into isolated mitochondria with equal efficiency over a concentration range of 8 to 70 nM. The experimentally determined in vivo concentration of the initiator tRNAMet variant is 63 nM (Crausaz-Esseiva et al, 2004a), and the one of the cytosolically localized elongator tRNAMet variant (assayed in Figure 6C) is 520 nM, respectively (Tan et al, 2002b; Crausaz-Esseiva et al, 2004a). In vitro import of the in vivo cytosolically localized tRNAMet variant is therefore not due to an unphysiological high substrate concentration.
Figure 6.
In vitro import assays. (A) Immunoblot analysis of 25 mg each of total cellular extract (Tot) and purified mitochondria (Mit) used in the in vitro assays for the presence of eEF1a (EF, top panel) and the mitochondrial marker α-ketoglutarate dehydrogenase (KDH, bottom panel), respectively (B) Left and middle panels: In vitro import in the presence and absence of ATP of in vitro transcribed and radioactively labeled imported elongator tRNAMet or cytosol-specific initiator tRNAMet, respectively. The input lanes depict 1% of the added substrate. Right panel: In vitro import of in vitro transcribed cytosol-specific tRNASec. In this case the imported tRNASec was detected by Northern blot and specific oligonucleotide hybridization. The concentration of each substrate tRNA in the import reaction is indicated. (C) In vitro import assays containing ATP and the indicated concentrations of in vitro transcribed tagged in vivo in part imported elongator tRNAMet (elongator tRNAMet*, top panel) or an in vivo cytosol-specific variant thereof carrying the T-arm of the initiator tRNAMet (elongator tRNAMet*T−arm Met−i, bottom panel). Imported tRNAs were detected by Northern blots and hybridization of oligonucleotides directed against the tag (Crausaz-Esseiva et al, 2004a). The input lanes show the hybridization signals obtained by 1 ng of substrate. The graph shows the quantification of the Northern blots. All import reactions shown in panels B and C were treated with micrococcus nuclease. Ethidium bromide-stained panels show the two mitochondrial rRNAs and serve as loading controls.
In summary these results suggest that the specificity of tRNA import is mediated by a cytosolic factor that is absent from the in vitro assay, and thus support our model.
Absence of eEF1a leads to unspecific import in vitro, but interferes with the membrane translocation step in vivo. This indicates that in living cells the targeting step is obligatory for the subsequent membrane translocation, whereas in the in vitro assay this step can be bypassed.
Discussion
The only cytosol-specific tRNAs in T. brucei are the initiator tRNAMet (Crausaz-Esseiva et al, 2004a) and the tRNASec (Geslain et al, 2006). Both of these tRNAs would be of no use inside the organelle, since the mitochondrial translation initiation mechanism is very different from the eukaryotic one (Tan et al, 2002a), and since no selenocysteine insertion machinery exists in mitochondria. Here we show that the lack of interaction with eEF1a provides an explanation for the cytosolic localization of these tRNAs. There are five lines of evidence supporting this conclusion: (i) an extensive in vivo study showed a perfect correlation of mitochondrial import of tRNAs with their predicted binding to eEF1a (Crausaz-Esseiva et al, 2004a); (ii) the main cytosolic localization determinant in the initiator tRNAMet coincides with a predicted antideterminant for eEF1a binding (Drabkin et al, 1998; Crausaz-Esseiva et al, 2004a); (iii) ablation of eEF1a abolishes import of newly synthesized tRNAs; (iv) a variant of the tRNASec that, unlike its wild-type counterpart, is predicted to bind to eEF1a, is imported into mitochondria by an eEF1a-dependent pathway (Figure 5) and (v) in vitro import of tRNAs in an in vitro system lacking eEF1a does not show specificity (Figure 6).
tRNA import can be subdivided into two temporally and spatially ordered steps. These are targeting of a subset of cytosolic tRNAs for mitochondrial import and the actual membrane translocation step. The specific interaction of eEF1a with imported tRNAs and the fact that it is a cytosolic protein that is never imported into mitochondria, indicate that eEF1a is involved in the targeting step. Thus, we suggest that besides its canonical function in translation, eEF1a selects a subpopulation of cytosolic tRNAs and hands them over to a putative receptor on the outer membrane of mitochondria. The receptor itself cannot discriminate between cytosol-specific tRNAs and tRNAs destined to be imported, which explains the lack of specificity in the in vitro assay (Figure 6). However, the membrane receptor appears to be able to monitor the modification status of tRNAs, as evidenced by in vitro import assay using Leishmania mitochondria. In these experiments, it was shown that the tRNAGln and tRNAGlu carrying a thio-modified anticodon nucleotide are less efficiently imported than the corresponding tRNAs lacking it (Kaneko et al, 2003). Thus, while the specificity of tRNA import is controlled by cytosolic eEF1a, the extent of import might be mediated by the modification status of tRNAs.
Interestingly, unlike what one might expect, ablation of eEF1a does not selectively interfere with the targeting step, but also prevents the membrane translocation process (Figure 4). This suggests that in living cells, in contrast to the in vitro import assay, the targeting step is obligatory for the subsequent membrane translocation of tRNAs. In the cell line ablated for eEF1a, impairment of tRNA import occurs well before inhibition of cytosolic translation (Figure 4). This shows that determining the tRNA import specificity and translation elongation are separate functions most likely mediated by two distinct eEF1a populations. Thus, it is conceivable that a small fraction of trypanosomal eEF1a, instead of transferring the tRNA to the A-site of the ribosome, hands it over to a putative tRNA import receptor on the surface of the mitochondrion. It is at present not known whether this requires ongoing translation elongation or not. There is evidence that in eukaryotes protein synthesis is a channeled pathway, meaning that elongator aminoacyl-tRNAs are directly transferred from the aminoacyl-tRNA synthetases to eEF1a and then to the ribosomes (Stapulionis and Deutscher, 1995; Hudder et al, 2003). This would mean that free aminoacyl-tRNAs that are not bound to proteins might not exist in the cell. Thus, we propose that in vivo import requires a highly structured cytosolic translation machinery.
However, the protein free tRNAs, that are used as substrates in the in vitro import system, may directly interact with the putative import receptor and as a consequence bypass the requirement for eEF1a.
Mitochondrial tRNA import is widespread among eukaryotes. However, contrary to mitochondrial protein import it has a polyphyletic evolutionary origin (Schneider and Marechal-Drouard, 2000). Thus, the tRNA import machineries might be distinct in different organisms and are probably less complex than the conserved, multicomponent protein import apparatus. This raises the question whether the recruitment of eEF1a for mitochondrial tRNA targeting is a general phenomenon? The number of tRNAs that are imported in the different systems is highly variable. In plants, for example there is no correlation of predicted eEF1a binding of tRNAs with mitochondrial import. An involvement of eEF1a in determining the tRNA import specificity is therefore unlikely. Interestingly, however, the tRNA import specificity in apicomplexan parasites, such as Plasmodium falciparum and Toxoplasma gondii, is probably identical to the one in trypanosomatids (Crausaz-Esseiva et al, 2004b). Thus, the role of eEF1a in tRNA targeting might well be conserved between these two groups even though they are not obviously related.
While the targeting function of eEF1a is unlikely to be conserved in all systems that import tRNAs, it is clear that a cytosolic targeting mechanism is also operative in S. cerevisiae. Targeting of the imported tRNALys isoacceptor to the surface of mitochondria requires the glycolytic enzyme enolase (Entelis et al, 2006). Thus, comparable to eEF1a in T. brucei, enolase has acquired a second function. It recognizes the imported tRNALys isoacceptor and hands it over to the precursor of mitochondrial lysyl-tRNA synthetase, the carrier protein required for import.
Recruiting housekeeping components appears to be a general feature of mitochondrial tRNA import in all systems. tRNA import in yeast makes use of the protein import pathway (Tarassov and Martin, 1996). Recent studies on Leishmania have suggested that the alpha subunit of the F1-ATPase (Goswami et al, 2006) as well as subunit 6b of the ubiquinol cytochrome c reductase (Chatterjee et al, 2006) are involved in the membrane translocation of tRNAs. Finally, studies in plants have shown that the voltage-dependent anion channel of the outer mitochondrial membrane is a major component of the tRNA import machinery (Salinas et al, 2006). In fact up to now no factor has been described yet in any system that functions in mitochondrial tRNA import only.
A number of non-canonical functions of eEF1a have been described. It has long been known that eEF1a is an actin-binding protein. Recently, mutations in eEF1a have been produced that alter actin cytoskeleton organization without interfering with protein synthesis (Gross and Kinzy, 2005). In plants eEF1a appears to be implicated in microtubule bundling (Durso and Cyr, 1994). Furthermore, overexpression of eEF1a was proposed to be important for apoptosis (Lamberti et al, 2004). Our results show that in T. brucei eEF1a has yet another novel function, namely mediating the specificity of mitochondrial tRNA import.
Materials and methods
Inducible tRNA expression
The inducible tRNA expression system is based on the tetracycline-regulatable polymerase I promoter system originally developed by (Wirtz and Clayton, 1995). To establish the system we used a derivative of pLew-100 where the 2296-bp KpnI/BamHI fragment was replaced by a KpnI/BamHI fragment consisting of the 21-bp tetracycline operator followed by a variant initiator tRNAMet gene, followed by 77-bp 3′-flanking region of the wild-type initiator tRNAMet. The variant initiator tRNAMet gene that was used contains the T-stem loop region of the elongator tRNAMet and a tag (G12:C23 to U12:A23) in the D-stem. Previous work has shown that this variant tRNAMet is imported into mitochondria of transgenic T. brucei to the same level as wild-type elongator tRNAMet (Crausaz-Esseiva et al, 2004a). Furthermore, it was shown that the tagged tRNA can be aminoacylated in vitro and is correctly processed in vivo. The construct was linearized with NotI and electroporated into T. brucei 29–13 grown in SDM-79 supplemented with 15% FCS. Transformants were selected with phleomycine and cloned as previously described (Beverley and Clayton, 1993). The selected clone allowed tetracycline-inducible (1 μg/ml) polymerase III-directed transcription of the variant initiator tRNAMet that could specifically be detected using the oligonucleotide 5′CGCTCTT CCCCTGAGCCA3′, which hybridizes to the region containing the D-stem tag.
RNAi cell lines
RNAi of eEF1a and eIF2 was performed using stem loop constructs containing the puromycine resistance gene, as described (Bochud-Allemann and Schneider, 2002). As inserts we used a 546-bp fragment (nucleotides 321–867) of the eEF1a gene and a 624-bp fragment of the eIF2 gene (nucleotides 257–881) (Berriman et al, 2005). The NotI-linearized constructs were electroporated into the clonal cell line obtained above. Selection with puromycine resulted in two new clonal cell lines that allow concomitant inducible expression of the variant tRNA, as well as ablation of either eEF1a or eIF2, respectively.
Expression of variant tRNASec
DNA fragments consisting of the tRNASec gene, containing the changes indicated in Figure 5A or C, respectively, including 308 bp of its 5′-flanking and 205 bp of its 3′ flanking region, were cloned into a modified pLew-100 containing convenient cloning sites downstream of the procyclin promoter. Linearization, electroporation, selection with puromycine and cloning were performed as above. In the presence of 1 μg/ml tetracycline the resulting clonal cell line expresses both wild-type tRNASec, which can be monitored by hybridization with the oligonucleotide 5′ACCAGCTGAGCTCAT CGTGGC3′, as well as either of the variant tRNAsSec, which can be specifically detected by hybridization with the oligonucleotides 5′TGGCACCACCACGGCCGA3′ (var1-tRNASec) or 5′TGGGACCACC ACGGCCGA3′ (var2-tRNASec).
Cell fractionation
Mitochondrial fractions were prepared by digitonin extractions (Tan et al, 2002b). Washed cells (4 × 108 cells each) were resuspended in 0.5 ml of SoTE (0.6 M sorbitol, 20 mM Tris–HCl, pH 7.5 and 2 mM EDTA). Five percent of the sample (25 μl) was removed to isolate the total RNA using the acidic guanidinium isothiocyanate method (Chomczyinski and Sacchi, 1987). After the addition of 0.475 ml of SoTE containing 0.1% (w/v) of digitonin, the samples were mixed by pipetting and incubated on ice for 5 min. The suspension (final concentration of digitonin 0.25%) was centrifuged (8000 g/5 min/4°C) and the supernatants were discarded. Next, the resulting pellets were resuspended in 500 μl of SoTE containing 1 μg of RNase A and incubated on ice for 15 min. After a final centrifugation the supernatants were discarded and RNA was isolated as for the total RNA sample. Both total RNA (corresponding to 2 × 107 cell equivalents) and mitochondrial RNA (corresponding to 2 × 107 cell equivalents) were separated on short 8 M urea/10% polyacrylamide gels.
Nuclear and cytosolic RNA fractions were prepared by digitonin extractions as described above, except that a final concentration of 0.1% of digitonin was used, 8% (w/v) of poly-(N-vinyl pyrrolidone) was added and no RNase digestion was performed. The nuclear RNA was isolated from the pellet and the cytosolic RNA from the supernatant. RNAs corresponding to 0.8 × 107 cell equivalents (for the cytosol) and 3.2 × 107 cell equivalents (for the nuclear fraction) were analyzed. U6 RNA was used as a nuclear marker and mature tRNAs visualized by ethidium bromide as a cytosolic marker (Figure 5B and C).
35S-labeling of total cellular proteins
The two RNAi cell lines containing the inducible tRNA expression system were induced with 1 μg/ml tetracycline for the indicated time. Next the cells were washed in phosphate buffer (20 mM sodium phosphate buffer, pH 7.9, 20 mM glucose, 0.15 M NaCl) and resuspended in SDM-80 (Lamour et al, 2005) that lacks methionine. Subsequently 35S-labeled methionine (1175 Ci/mmol) was added to a final concentration of 4 mCi/ml. After incubation for 2 h at 27°C, aliquots of 5 × 107 cells were removed and washed in phosphate buffer. The resulting pellets were resuspended in standard SDS–gel sample buffer and resolved on a 10% SDS–polyacrylamide gel. The dried gel was exposed on a phosphorimager and the total signal obtained per lane of the induced samples was compared to that of a lane containing the uninduced control.
In vitro import assays
A standard in vitro import reaction was performed in 20 ml of SoTE containing 2 mM DTT, 20 mM MgCl2 and isotonically isolated mitochondria (500 μg protein) (Hauser et al, 1996; Schneider et al, 2007). After the addition of the indicated amounts of the different substrate tRNAs (Figure 6), the reaction was incubated for 20 min at 27°C in either the absence or the presence of a mixture containing 8 mM ATP, 1.3 mM creatine phosphate and 1 μg of creatine kinase (Roche). Subsequently, a tube was prepared containing a bottom layer of 10 ml of 20 mM Tris–HCl, pH 8.0, 2 mM EDTA containing 1.75 M of sucrose and a top layer of 20 μl of the same buffer containing 0.6 M of sucrose. The 20-μl import reaction was overlayed on the top layer and after centrifugation for 5 min (6800 g at 4°C) the top 30 μl were discarded. The remaining 20 μl were mixed by pipetting. Subsequently, CaCl2 was added to a final concentration of 2.2 mM and the reaction was digested with 48 U of micrococcal nuclease (MBI Fermentas) for 15 min at 4°C, followed by 30 min at 27°C. Finally, RNA was isolated using the guanidinium isothiocyanate method (Chomczyinski and Sacchi, 1987) and analyzed on a short 8 M urea/10% polyacrylamide sequencing gel.
Miscellaneous
Transfer and Northern hybridization using specific radioactively kinased oligonucleotide probes were performed as described (Tan et al, 2002b).
Supplementary Material
Supplementary Figure 1
Supplementary Figure Legends
Acknowledgments
We thank E Horn and L Bulliard for technical assistance, and also thank G Cross for providing us with cell lines and plasmids. This work was supported by grant 3100–067906 of the Swiss National Foundations and by a grant from the Velux Foundation.
References
- Baker KP, Schaniel A, Vestweber D, Schatz G (1990) A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature 348: 605–609 [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcell L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ et al. (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309: 416–422 [DOI] [PubMed] [Google Scholar]
- Beverley SM, Clayton CE (1993) Transfection of Leishmania and Trypanosoma brucei by electroporation. Methods Mol Biol 21: 333–348 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Adhya S (2004) The complexity of mitochondrial tRNA import. RNA Biol 1: 84–88 [DOI] [PubMed] [Google Scholar]
- Bochud-Allemann N, Schneider A (2002) Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J Biol Chem 277: 32849–32854 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21: 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Treichel N, Muller EC, Otto A, Kutay U (2002) Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 21: 6216–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassago A, Rodrigues EM, Prieto EL, Gaston KW, Alfonzo JD, Iribar MP, Berry MJ, Cruz AK, Thiemann OH (2006) Identification of Leishmania selenoproteins and SECIS element. Mol Biochem Parasitol 149: 128–134 [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Home P, Mukherjee S, Mahata B, Goswami S, Dhar G, Adhya S (2006) An RNA-binding respiratory component mediates import of type II tRNAs into Leishmania mitochondria. J Biol Chem 281: 2570–2577 [DOI] [PubMed] [Google Scholar]
- Chomczyinski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Crausaz-Esseiva A, Marechal-Drouard L, Cosset A, Schneider A (2004a) The T-stem determines the cytosolic or mitochondrial localization of trypanosomal methionyl-tRNAs. Mol Biol Cell 15: 2750–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crausaz-Esseiva A, Naguleswaran A, Hemphill A, Schneider A (2004b) Mitochondrial tRNA import in Toxoplasma gondii. J Biol Chem 279: 42363–42368 [DOI] [PubMed] [Google Scholar]
- Delage L, Duchene AM, Zaepfel M, Marechal-Drouard L (2003) The anticodon and the D-domain sequences are essential determinants for plant cytosolic valyl-tRNA import into mitochondria. Plant J 34: 623–633 [DOI] [PubMed] [Google Scholar]
- Diamond AM (2004) On the road to selenocysteine. Proc Natl Acad Sci USA 101: 133395–133396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich A, Marechal-Drouard L, Carneiro V, Cosset A, Small I (1996a) A single base change prevents import of cytosolic analyl-tRNA into mitochondria in transgenic plants. Plant J 10: 913–918 [DOI] [PubMed] [Google Scholar]
- Dietrich A, Small I, Cosset A, Weil JH, Marechal-Drouard L (1996b) Editing and import: strategies for providing plant mitochondria with a complete set of functional transfer RNAs. Biochimie 78: 502–510 [DOI] [PubMed] [Google Scholar]
- Drabkin HJ, Estrella M, Rajbhandary UL (1998) Initiator–elongator discrimination in vertebrate tRNAs for protein synthesis. Mol Cell Biol 18: 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Uhlenbeck OC, Browning KS (1999) Quantitative assessment of EF-1alpha. GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J Biol Chem 274: 666–672 [DOI] [PubMed] [Google Scholar]
- Durso NA, Cyr RJ (1994) A calmodulin-sensitive interaction between microtubules and a higher plant homolog of elongation factor-1 alpha. Plant Cell 6: 893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis N, Brandina I, Kamenski P, Krasheninnikov IA, Martin RP, Tarassov I (2006) A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev 20: 1609–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entelis NS, Kiefer S, Kolesnikova OA, Martin RP, Tarassov IA (1998) Structural requirements of lysyl-tRNA for its import into yeast mitochondria. Proc Natl Acad Sci USA 95: 2838–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geslain R, Aeby E, Guitart T, Jones TE, Moura MCd, Charrière F, Schneider A, Pouplana LRd (2006) Trypanosoma seryl-tRNA synthetase is a metazoan-like enzyme with high affinity for tRNASec. J Biol Chem 281: 38217–38225 [DOI] [PubMed] [Google Scholar]
- Goswami S, Dhar G, Mukherjee S, Mahata B, Chatterjee S, Home P, Adhya S (2006) A bifunctional tRNA import receptor from Leishmania mitochondria. Proc Natl Acad Sci USA 103: 8354–8359 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gross SR, Kinzy TG (2005) Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat Struct Mol Biol 9: 772–778 [DOI] [PubMed] [Google Scholar]
- Hancock K, Hajduk SL (1990) The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J Biol Chem 265: 19208–19215 [PubMed] [Google Scholar]
- Hauser R, Pypaert M, Häusler T, Horn EK, Schneider A (1996) In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J Cell Sci 109: 517–523 [DOI] [PubMed] [Google Scholar]
- Hudder A, Nathanson L, Deutscher MP (2003) Organization of mammalian cytoplasm. Mol Cell Biol 23: 9318–9326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Suzuki T, Kapushoc ST, Rubio MA, Ghazvini J, Watanabe K, Simpson L, Suzuki T (2003) Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism. EMBO J 22: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur KJ, Ruben L (1994) Protein translation elongation factor-1alpha from Trypanosoma brucei binds calmodulin. J Biol Chem 269: 23045–23050 [PubMed] [Google Scholar]
- Lamberti A, Caraglia M, Longo O, Marra M, Abbruzzese A, Arcari P (2004) The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids 26: 443–448 [DOI] [PubMed] [Google Scholar]
- Lamour N, Riviere L, Coustou V, Coombs GH, Barrett MP, Bringaud F (2005) Proline metabolism in procyclic Trypanosoma brucei is downregulated in the presence of glucose. J Biol Chem 280: 11902–11910 [DOI] [PubMed] [Google Scholar]
- Lobanov AV, Gromer S, Salinas G, Gladyshev VN (2006) Selenium metabolism in Trypanosoma: characterization of selenoproteomes and identification of a Kinetoplastida-specific selenoprotein. Nucleic Acids Res 34: 4012–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Bhattacharyya SN, Adhya S (1999) Stepwise transfer of tRNA through the double membrane of Leishmania mitochondria. J Biol Chem 274: 31249–31255 [DOI] [PubMed] [Google Scholar]
- Ribeiro S, Nock S, Sprinzl M (1995) Purification of aminoacyl-tRNA by affinity chromatography on immobilized Thermus thermophilus EF-Tu.GTP. Anal Biochem 228: 330–335 [DOI] [PubMed] [Google Scholar]
- Rinehart J, Krett B, Rubio MA, Alfonzo JD, Soll D (2005) Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev 19: 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MA, Liu X, Yuzawa H, Alfonzo JD, Simpson L (2000) Selective importation of RNA into isolated mitochondria from Leishmania tarentolae RNA. RNA 6: 988–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger J, Hillenbrandt R, Sprinzl M, Giege R (1996) Antideterminants present in minihelix(Sec) hinder its recognition by prokaryotic elongation factor Tu. EMBO J 15: 650–657 [PMC free article] [PubMed] [Google Scholar]
- Rusconi CP, Cech TR (1996) The anticodon is the signal sequence for mitochondrial import of glutamine tRNA in Tetrahymena. Genes Dev 10: 2870–2880 [DOI] [PubMed] [Google Scholar]
- Salinas T, Duchene AM, Delage L, Nilsson S, Glaser E, Zaepfel M, Marechal-Drouard L (2006) The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc Natl Acad Sci USA 103: 18362–18367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Charrière F, Pusnik M, Horn EK (2007) Isolation of mitochondria from procyclic Trypanosoma brucei. Methods Mol Biol 372: 67–80 [DOI] [PubMed] [Google Scholar]
- Schneider A, Marechal-Drouard L (2000) Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell Biol 10: 509–513 [DOI] [PubMed] [Google Scholar]
- Schneider A, Martin JA, Agabian N (1994) A nuclear encoded tRNA of Trypanosoma brucei is imported into mitochondria. Mol Cell Biol 14: 2317–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AM, Suyama Y, Dewes H, Campbell DA, Simpson L (1989) Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res 17: 5427–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapulionis R, Deutscher MP (1995) A channeled tRNA cycle during mammalian protein synthesis. Proc Natl Acad Sci USA 92: 7158–7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan THP, Bochud-Allemannn N, Horn EK, Schneider A (2002a) Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria. Proc Natl Acad Sci USA 99: 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan THP, Pach R, Crausaz A, Ivens A, Schneider A (2002b) tRNAs in Trypanosoma brucei: genomic organization, expression and mitochondrial import. Mol Cell Biol 22: 3707–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov I, Entelis N, Martin RP (1995) Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J 14: 3461–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov I, Martin R (1996) Mechanisms of tRNA import into yeast mitochondria: an overview. Biochimie 78: 502–510 [DOI] [PubMed] [Google Scholar]
- Wirtz E, Clayton C (1995) Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science 268: 1179–1183 [DOI] [PubMed] [Google Scholar]
- Yermovsky-Kammerer AE, Hajduk S (1999) In vitro import of a nuclearly encoded tRNA into the mitochondrion of Trypanosoma brucei. Mol Cell Biol 19: 6253–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure Legends